Sequencing - PowerPoint PPT Presentation
1 / 71
Title:
Sequencing
Description:
Be aware of the importance of sequence searching and sequence alignment in biology and medicine ... every 20 residues the alignment is suspicious. 29. Lecture ... – PowerPoint PPT presentation
Number of Views:350
Avg rating:3.0/5.0
Title: Sequencing
1
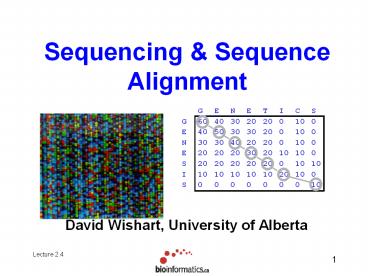
Sequencing Sequence Alignment
David Wishart, University of Alberta
2
Objectives
- Understand how DNA sequence data is collected and
prepared - Be aware of the importance of sequence searching
and sequence alignment in biology and medicine - Be familiar with the different algorithms and
scoring schemes used in sequence searching and
sequence alignment
3
High Throughput DNA Sequencing
4
30,000
5
Shotgun Sequencing
Isolate Chromosome
ShearDNA into Fragments
Clone into Seq. Vectors
Sequence
6
Principles of DNA Sequencing
Primer
DNA fragment
Amp
PBR322
Tet
Ori
Denature with heat to produce ssDNA
Klenow ddNTP dNTP primers
7
The Secret to Sanger Sequencing
8
Principles of DNA Sequencing
3 Template
G C A T G C
5
5 Primer
GddC
GCddA
GCAddT
ddG
GCATGddC
GCATddG
9
Principles of DNA Sequencing
G
T
_
_
C
A
G C A T G C
10
Capillary Electrophoresis
Separation by Electro-osmotic Flow
11
Multiplexed CE with Fluorescent detection
ABI 3700
96x700 bases
12
Shotgun Sequencing
Assembled Sequence
Sequence Chromatogram
Send to Computer
13
Shotgun Sequencing
- Very efficient process for small-scale (10 kb)
sequencing (preferred method) - First applied to whole genome sequencing in 1995
(H. influenzae) - Now standard for all prokaryotic genome
sequencing projects - Successfully applied to D. melanogaster
- Moderately successful for H. sapiens
14
The Finished Product
GATTACAGATTACAGATTACAGATTACAGATTACAG ATTACAGATTACA
GATTACAGATTACAGATTACAGA TTACAGATTACAGATTACAGATTACA
GATTACAGAT TACAGATTAGAGATTACAGATTACAGATTACAGATT AC
AGATTACAGATTACAGATTACAGATTACAGATTA CAGATTACAGATTAC
AGATTACAGATTACAGATTAC AGATTACAGATTACAGATTACAGATTAC
AGATTACA GATTACAGATTACAGATTACAGATTACAGATTACAG ATTA
CAGATTACAGATTACAGATTACAGATTACAGA TTACAGATTACAGATTA
CAGATTACAGATTACAGAT
15
Sequencing Successes
T7 bacteriophage completed in 1983 39,937 bp, 59
coded proteins Escherichia coli completed in
1998 4,639,221 bp, 4293 ORFs Sacchoromyces
cerevisae completed in 1996 12,069,252 bp, 5800
genes
16
Sequencing Successes
Caenorhabditis elegans completed in
1998 95,078,296 bp, 19,099 genes Drosophila
melanogaster completed in 2000 116,117,226 bp,
13,601 genes Homo sapiens 1st draft completed in
2001 3,160,079,000 bp, 31,780 genes
17
So what do we do with all this sequence data?
18
Sequence Alignment
19
Alignments tell us about...
- Function or activity of a new gene/protein
- Structure or shape of a new protein
- Location or preferred location of a protein
- Stability of a gene or protein
- Origin of a gene or protein
- Origin or phylogeny of an organelle
- Origin or phylogeny of an organism
20
Factoid
Sequence comparisons lie at the heart of
all bioinformatics
21
Similarity versus Homology
- Similarity refers to the likeness or identity
between 2 sequences - Similarity means sharing a statistically
significant number of bases or amino acids - Similarity does not imply homology
- Homology refers to shared ancestry
- Two sequences are homologous is they are derived
from a common ancestral sequence - Homology usually implies similarity
22
Similarity versus Homology
- Similarity can be quantified
- It is correct to say that two sequences are X
identical - It is correct to say that two sequences have a
similarity score of Z - It is generally incorrect to say that two
sequences are X similar
23
Similarity versus Homology
- Homology cannot be quantified
- If two sequences have a high identity it is OK
to say they are homologous - It is incorrect to say two sequences have a
homology score of Z - It is incorrect to say two sequences are X
homologous
24
Sequence Complexity
MCDEFGHIKLAN. High Complexity
ACTGTCACTGAT. Mid Complexity
NNNNTTTTTNNN. Low Complexity
Translate those DNA sequences!!!
25
Assessing Sequence Similarity
THESTORYOFGENESIS THISBOOKONGENETICS THESTORYOFGE
NESI-S THISBOOKONGENETICS THE STORY OF
GENESIS THIS BOOK ON GENETICS
Two Character Strings
Character Comparison
Context Comparison
26
Assessing Sequence Similarity
is this alignment significant?
27
Is This Alignment Significant?
28
Some Simple Rules
- If two sequence are gt 100 residues and gt
25 identical, they are likely related - If two sequences are 15-25 identical they may be
related, but more tests are needed - If two sequences are lt 15 identical they are
probably not related - If you need more than 1 gap for every 20 residues
the alignment is suspicious
29
Doolittles Rules of Thumb
30
Sequence Alignment - Methods
- Dot Plots
- Dynamic Programming
- Heuristic (Fast) Local Alignment
- Multiple Sequence Alignment
- Contig Assembly
31
Dot Plots
32
Dot Plots
- Invented in 1970 by Gibbs McIntyre
- Good for quick graphical overview
- Simplest method for sequence comparison
- Inter-sequence comparison
- Intra-sequence comparison
- Identifies internal repeats
- Identifies domains or modules
33
Dot Plots Internal Repeats
34
Dot Plot Algorithm
- Take two sequences (A B), write sequence A out
as a row (lengthm) and sequence B as a column
(length n) - Create a table or matrix of m columns and n
rows - Compare each letter of sequence A with every
letter in sequence B. If theres a match mark it
with a dot, if not, leave blank
35
Dot Plot Algorithm
A C D E F G H G
A C D E F G H G
36
Dot Plots
- Most commercial programs offer pretty good dot
plot programs including - GCG/Omiga (Pharmacopeia)
- PepTool (BioTools Inc.)
- LaserGene (DNAStar)
- Popular freeware package is Dotter
www.cgr.ki.se/cgr/groups/sonnhammer/Dotter.html - Dotlet http//www.isrec.isb-sib.ch/java/dotlet/Dot
let.html - JDotter http//athena.bioc.uvic.ca/sars/jdotter/ma
in.php
37
Dynamic Programming
38
Dynamic Programming
- Developed by Needleman Wunsch (1970)
- Refined by Smith Waterman (1981)
- Ideal for quantitative assessment
- Guaranteed to be mathematically optimal
- Slow N2 algorithm
- Performed in 2 stages
- Prepare a scoring matrix using recursive function
- Scan matrix diagonally using traceback protocol
39
The Recursive Function
Si-1,j-1 or max Si-x,j-1 wx-1
or max Si-1,j-y wy-1
Sij sij max
2ltxlti
2ltyltj
W gap penalty S alignment score
40
Identity Scoring Matrix (Sij)
41
A Simple Example...
A A T V D A 1 V V D
A A T V D A 1 1 V V D
A A T V D A 1 1 0 0 0 V V D
A A T V D A 1 1 0 0 0 V 0 V D
A A T V D A 1 1 0 0 0 V 0 1 1 V D
A A T V D A 1 1 0 0 0 V 0 1 1 2 V D
42
A Simple Example...
A A T V D A 1 1 0 0 0 V 0 1 1 2 1 V D
A A T V D A 1 1 0 0 0 V 0 1 1 2 1 V 0 1
1 2 2 D 0 1 1 1 3
A A T V D A 1 1 0 0 0 V 0 1 1 2 1 V 0 1
1 2 2 D 0 1 1 1 3
A A T V D A - V V D
A A T V D A V V D
A A T V D A V - V D
43
Could We Do Better?
- Key to the performance of Dynamic Programming is
the scoring function - Dynamic Programming always gives the
mathematically correct answer - Dynamic Programming does not always give the
biologically correct answer - The weakest link -- The Scoring Matrix
44
Scoring Matrices
- An empirical model of evolution, biology and
chemistry all wrapped up in a 20 X 20 table of
integers - Structurally or chemically similar residues
should ideally have high diagonal or off-diagonal
numbers - Structurally or chemically dissimilar residues
should ideally have low diagonal or off-diagonal
numbers
45
A Better Matrix - PAM250
46
Using PAM250...
A T V D A 2 T 1 3 V 0 0 4 D 0 0-2 4
Gap Penalty -1
A A T V D A 2 V V D
A A T V D A 2 1 V V D
A A T V D A 2 1 0 -1 -1 V V D
A A T V D A 2 1 0 -1 -1 V -1 2 V D
A A T V D A 2 1 0 -1 -1 V -1 2 1
V D
A A T V D A 2 1 0 -1 -1 V -1 2 1 5 V D
47
Using PAM250...
A T V D A 2 T 1 3 V 0 0 4 D 0 0-2 4
Gap Penalty -1
A A T V D A 2 1 0 -1 -1 V -1 2 1 5
-1 V D
A A T V D A 2 1 0 -1 -1 V -1 2 1 5 -1 V
-1 1 2 5 3 D -1 1 1 0 9
A A T V D A 2 1 0 -1 -1 V -1 2 1 5 -1 V
-1 1 2 5 3 D -1 1 1 0 9
A A T V D A V - V D
48
PAM Matrices
- Developed by M.O. Dayhoff (1978)
- PAM Point Accepted Mutation
- Matrix assembled by looking at patterns of
substitutions in closely related proteins - 1 PAM corresponds to 1 amino acid change per 100
residues - 1 PAM 1 divergence or 1 million years in
evolutionary history
49
Dynamic Programming
- Great for doing pairwise global alignments
- Produces a quantitative alignment score
- Problems if one tries to do alignments with very
large sequences (memory requirement grows as N2
or as N x M) - Serious problems if one tries to align one
sequence against a database (10s of hours) - Need an alternative..
50
Fast Local Alignment Methods
ACDEAGHNKLM...
KKDEFGHPKLM...
SCDEFCHLKLM...
MCDEFGHNKLV...
ACDEFGHIKLM...
QCDEFGHAKLM...
AQQQFGHIKLPI...
WCDEFGHLKLM...
SMDEFAHVKLM...
ACDEFGFKKLM...
51
Fast Local Alignment Methods
- Developed by Lipman Pearson (1985/88)
- Refined by Altschul et al. (1990/97)
- Ideal for large database comparisons
- Uses heuristics statistical simplification
- Fast N-type algorithm (similar to Dot Plot)
- Cuts sequences into short words (k-tuples)
- Uses Hash Tables to speed comparison
52
Fast Alignment Algorithm
53
Fast Alignment Algorithm
54
Fast Alignment Algorithm
A C D E F G D E F...
L M R G CD D Y G
55
Fast Alignment Algorithm
56
FASTA
- Developed in 1985 and 1988 (W. Pearson)
- Looks for clusters of nearby or locally dense
identical k-tuples - init1 score score for first set of k-tuples
- initn score score for gapped k-tuples
- opt score optimized alignment score
- Z-score number of S.D. above random
- expect expected of random matches
57
FASTA
58
Multiple Sequence Alignment
Multiple alignment of Calcitonins
59
Multiple Alignment Algorithm
- Take all n sequences and perform all possible
pairwise (n/2(n-1)) alignments - Identify highest scoring pair, perform an
alignment create a consensus sequence - Select next most similar sequence and align it to
the initial consensus, regenerate a second
consensus - Repeat step 3 until finished
60
Multiple Sequence Alignment
- Developed and refined by many (Doolittle, Barton,
Corpet) through the 1980s - Used extensively for extracting hidden
phylogenetic relationships and identifying
sequence families - Powerful tool for extracting new sequence motifs
and signature sequences
61
Multiple Alignment
- Most commercial vendors offer good multiple
alignment programs including - GCG (Accelerys)
- PepTool/GeneTool (BioTools Inc.)
- LaserGene (DNAStar)
- Popular web servers include T-COFFEE, MULTALIN
and CLUSTALW - Popular freeware includes PHYLIP PAUP
62
Mutli-Align Websites
- Match-Box http//www.fundp.ac.be/sciences/biologie
/bms/matchbox_submit.shtml - MUSCA http//cbcsrv.watson.ibm.com/Tmsa.html
- T-Coffee http//www.ch.embnet.org/software/TCoffee
.html - MULTALIN http//www.toulouse.inra.fr/multalin.html
- CLUSTALW http//www.ebi.ac.uk/clustalw/
63
Multi-alignment Contig Assembly
ATCGATGCGTAGCAGACTACCGTTACGATGCCTT TAGCTACGCATCGT
CTGATGGCAATGCTACGGAA..
TAGCTACGCATCGT
TAGCAGACTACCGTT
ATCGATGCGTAGC
GTTACGATGCCTT
64
Contig Assembly
- Read, edit trim DNA chromatograms
- Remove overlaps ambiguous calls
- Read in all sequence files (10-10,000)
- Reverse complement all sequences (doubles of
sequences to align) - Remove vector sequences (vector trim)
- Remove regions of low complexity
- Perform multiple sequence alignment
65
Chromatogram Editing
66
Sequence Loading
67
Sequence Alignment
68
Contig Alignment - Process
ATCGATGCGTAGC
TAGCAGACTACCGTT
GTTACGATGCCTT
TGCTACGCATCG
CGATGCGTAGCA
CGATGCGTAGCA
ATCGATGCGTAGC
TAGCAGACTACCGTT
GTTACGATGCCTT
ATCGATGCGTAGCAGACTACCGTTACGATGCCTT
69
Sequence Assembly Programs
- Phred - base calling program that does detailed
statistical analysis (UNIX)
http//www.phrap.org/ - Phrap - sequence assembly program (UNIX)
http//www.phrap.org/ - TIGR Assembler - microbial genomes (UNIX)
http//www.tigr.org/softlab/assembler/ - The Staden Package (UNIX)
- http//www.mrc-lmb.cam.ac.uk/pubseq/
- GeneTool/ChromaTool/Sequencher (PC/Mac)
70
http//bio.ifom-firc.it/ASSEMBLY/assemble.html
71
Conclusions
- Sequence alignments and database searching are
key to all of bioinformatics - There are four different methods for doing
sequence comparisons 1) Dot Plots 2) Dynamic
Programming 3) Fast Alignment and 4) Multiple
Alignment - Understanding the significance of alignments
requires an understanding of statistics and
distributions