Protein Degradation In the Eukaryotic Cytoplasm - PowerPoint PPT Presentation
1 / 30
Title:
Protein Degradation In the Eukaryotic Cytoplasm
Description:
and rings each have seven subunits ... at the same time, or that an internal loop can be the initial engagement site ... RING domain E3's transfer ubiquitin ... – PowerPoint PPT presentation
Number of Views:348
Avg rating:3.0/5.0
Title: Protein Degradation In the Eukaryotic Cytoplasm
1
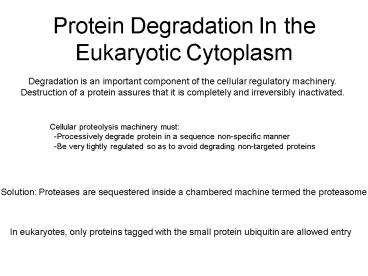
Protein Degradation In the Eukaryotic Cytoplasm
Degradation is an important component of the
cellular regulatory machinery. Destruction of a
protein assures that it is completely and
irreversibly inactivated.
Cellular proteolysis machinery must
-Processively degrade protein in a sequence
non-specific manner -Be very tightly regulated
so as to avoid degrading non-targeted proteins
Solution Proteases are sequestered inside a
chambered machine termed the proteasome
In eukaryotes, only proteins tagged with the
small protein ubiquitin are allowed entry
2
ATP-dependent Covalent Linkage to Ubiquitin and
Proteolysis
Labeled Ub
Labeled lysozyme
ATP-dependent covalent linkage of ubiquitin to
lysozyme
Conjugated protein subsequently degraded in
cell lysate
Varshavsky and colleagues later showed that
mutation of ubiquitin pathway blocks degradation
of many normal proteins
Work shown here and related work earned
authors the Nobel prize in Chemistry in 2004
Hershko et al, PNAS (1980) 77, 1783-1786.
3
Protein Degradation In the Eukaryotic Cytoplasm
1. Structures and properties of the proteasome
2. Tagging proteins for destruction by attachment
of ubiquitin
3. What determines which proteins are
ubiquitylated The N-end rule
4
Chambered Proteases In Archaea, Prokaryotes and
Eukaryotes
Pickart and Cohen, Nat. Rev. Mol. Cell Biol.
(2004) 5, 177-187
5
Electron-microscopy Images of Yeast Proteasome
? and ? rings each have seven subunits
? subunits control passage of polypeptide
substrates into ? region and may allow repeated
cycles of action
Three of seven ? subunits have protease active
sites, and there are two ? rings per proteasome
complex.
The entire proteasome therefore has six active
sites of three different types
Lid and base additions at end contain ATPases
Pickart and Cohen, Nat. Rev. Mol. Cell Biol.
(2004) 5, 177-187
6
Model of Eukaryotic Proteasome
Zwickl and Baumeister, Ann. Rev. Biochem. (1999)
68, 1015-1068
7
Subunit Composition of the Yeast Proteasome
ATPases
Proteases
Pickart and Cohen, Nat. Rev. Mol. Cell Biol.
(2004) 5, 177-187
8
Substrate Proteins Are Unfolded As They Enter the
Proteasome
Very large changes in substrate protein
stability give only small changes in degradation
rate
ATPases in 19S portion actively unfold proteins
to allow them to enter the proteasome
Mechanism is apparently to push or pull the
polypeptide chain into the cavity. Entry rate is
sensitive to structural features at initial
engagement site but not global stability
Peptides with intra-chain crosslinks can be
degraded, indicating that more than one
polypeptide chain can enter at the same time, or
that an internal loop can be the initial
engagement site
Once inside, the protein is extensively
degraded. The active sites hydrolyze peptide
bonds with high efficiency and little
specificity. Fragments are presumably trapped by
the lids until they are cleaved many times and
are very short.
Pickart and Cohen, Nat. Rev. Mol. Cell Biol.
(2004) 5, 177-187
9
Gate Opening in the Archaeal 20S Proteasome
Rabi et al, Mol. Cell (2008) 30, 360-368.
10
Gate Opening by Interaction of PAN Subunits
C-terminii With 20S Proteasome
Smith et al., Mol. Cell (2007) 27, 731-744.
11
Binding to Peptides From PAN Induces Gate Opening
At least one other lid complex is present in
archaea, which opens gate using a different
mechanism
Rabi et al, Mol. Cell (2008) 30, 360-368.
12
Protein Degradation In the Eukaryotic Cytoplasm
1. Structures and properties of the proteasome
2. Tagging proteins for destruction by attachment
of ubiquitin
3. What determines which proteins are
ubiquitylated The N-end rule
13
Pathway For Ubiquitination and Degradation By the
Proteasome
Zwickl and Baumeister, Ann. Rev. Biochem. (1999)
68, 1015-1068
14
Activation of Ubiquitination and Attachment to a
Target Protein
Ubiquitin is first activated by reaction with
ATP, catalyzed by E1 enzyme
Ubiquitin is then attached to E1 with a
thio-ester linkage, with AMP as leaving group
In the next step, ubiquitin is transferred to
E2, again with thio-ester linkage
Last, ubiquitin is transferred to Lys of target
protein in a process directed by E3
Pickart, Cell (2004) 116, 181-190
15
Activation of Ubiquitination and Attachment to a
Target Protein
Ubiquitin is first activated by reaction with
ATP, catalyzed by E1 enzyme
Ubiquitin is then attached to E1 with a
thio-ester linkage, with AMP as leaving group
In the next step, ubiquitin is transferred to
E2, again with thio-ester linkage
Last, ubiquitin is transferred to Lys of target
protein in a process directed by E3
Pickart, Cell (2004) 116, 181-190
16
Two Major Classes of E3 Enzyme
HECT domain E3s have intermediate in which
ubituitin is covalently linked to E3 RING domain
E3s transfer ubiquitin directly from E2 to
substrate
Pickart, Cell (2004) 116, 181-190
17
General Model For Targeting Ubiquitinated
Substrates to the Proteasome
Or the Ub chain is bound directly by Rpn10 within
the proteasome
Elasser and Finley, Nat. Cell Biol (2005) 7,
742-749
18
ATP Is Used in Two Steps of the Pathway For
Destruction of Protein
ATP is necessary both for ubiquitination
process and for unfolding of substrate proteins
Pickart, Cell (2004) 116, 181-190
19
While the Protein Substrate Is Destroyed,
Ubiquitin Is Spared
Ubiquitin is released, but only after substrate
is engaged and translocation begins
Rpn11 mediates ubiquitin release by breaking
its covalent connection with the substrate
protein. In 26S proteasome, the reaction is
strictly dependent on ATP. ATP-dependent
translocation is apparently necessary to
translocate the polypeptide into position for
removal of the ubiquitin
Substrates that contain a non-cleavable
ubiquitin variant G76V are ultimately
proteolyzed, including the ubiquitin, but
cleavage is much slower
Pickart and Cohen, Nat. Rev. Mol. Cell Biol.
(2004) 5, 177-187
20
Protein Degradation In the Eukaryotic Cytoplasm
1. Structures and properties of the proteasome
2. Tagging proteins for destruction by attachment
of ubiquitin
3. What determines which proteins are
ubiquitylated The N-end rule
21
What Targets a Protein For Destruction? The N-end
Rule
Initially discovered by A. Varshavsky in 1986
Varshavsky, PNAS (1996) 93, 12142-9
22
Experiments and Model of the N-end Rule
E3
Varshavsky, PNAS (1996) 93, 12142-9
23
Amino Acids Are Classified As Stabilizing or
Destabilizing
secondary
tertiary
Varshavsky, PNAS (1996) 93, 12142-9
24
What Targets a Protein For Destruction? The N-end
Rule
Secondary and tertiary destabilizing residues are
enzymatically converted into primary ones
Varshavsky, PNAS (1996) 93, 12142-9
25
Covalent Modifications Can Enhance Substrate
Recognition by E3 Enzymes
Some E3 enzymes bind hydrophobic amino acids,
apparently to recognize misfolded proteins
Pickart, Cell (2004) 116, 181-190
26
Key Points
1. Eukaryotic proteins are degraded in the
cytoplasm by a large, complex machine called the
proteasome. Proteins are protected from random
encounters with the proteasome by being denied
entry into the soluble inside chamber, where the
protease active sites are sequestered.
2. The process of degrading proteins depends on
ATP, both for tagging the proteins with ubiquitin
and for unfolding the proteins as they enter the
proteasome.
3. The N-terminal amino acid of proteins is an
important determinant of their susceptibility to
degradation by the proteasome.
27
(No Transcript)
28
(No Transcript)
29
(No Transcript)
30
(No Transcript)