Genome-scale constraint-based metabolic model of - PowerPoint PPT Presentation
Title:
Genome-scale constraint-based metabolic model of
Description:
Genome-scale constraint-based metabolic model of Clostridium thermocellum School of Engineering Chris M. Gowen1,3, Seth B. Roberts1, Stephen S. Fong1,2 – PowerPoint PPT presentation
Number of Views:59
Avg rating:3.0/5.0
Title: Genome-scale constraint-based metabolic model of
1
Genome-scale constraint-based metabolic model of
Clostridium thermocellum
Chris M. Gowen1,3, Seth B. Roberts1, Stephen S.
Fong1,2 1Department of Chemical and Life Science
Engineering, Virginia Commonwealth University,
Richmond, VA, USA 2Center for the Study of
Biological Complexity, Virginia Commonwealth
University, Richmond, VA, USA 3Presenting author,
contact gowencm_at_vcu.edu
Motivation Cellulose makes up roughly 60 of the
dry weight of all plant biomass on earth and
therefore represents an extremely abundant and
sustainable feedstock for the production of
liquid fuels. All current methods for the
biochemical conversion of cellulosic biomass to
ethanol for fuel fall into three main categories
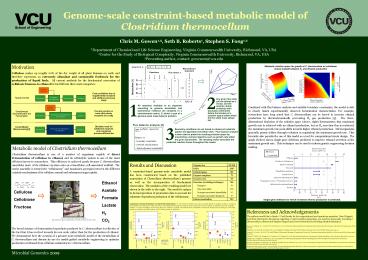
Metabolic solution space for growth of C.
thermocellum on cellobiose shows tradeoff between
H2 and ethanol production
2 A B ? C D
Reaction A
Mass balance statement
Reactions
Flux vector, v
A B C D
0 0 0 0 0 0
v1 . . . . .
Metabolites (X)
S v
dX / dt
Combined with flux balance analysis and suitable
boundary constraints, the model is able to
closely match experimentally observed
fermentation characteristics. For example,
researchers have long noted that C. thermocellum
can be forced to increase ethanol production by
thermodynamically preventing H2 gas production
3. The three-dimensional depiction of the
solution space (above, right) demonstrates that
maximum growth rate is achieved with no ethanol
production, but as H2 secretion flux is
restricted, the maximum growth rate peak shifts
towards higher ethanol production.
Microorganisms generally pursue (either through
evolution or regulation) the maximum growth rate.
This heuristic also permits the use of this
model as a tool for computational strain design.
The graph (below) shows single-gene deletions
predicted to improve ethanol secretion at the
maximum growth rate. This technique can be used
to inform genetic engineering decision making.
Flux balance analysis 2
Metabolic model of Clostridium thermocellum Clostr
idium thermocellum is one of a number of
organisms capable of direct fermentation of
cellulose to ethanol, and its cellulolytic system
is one of the most efficient known to
researchers. This efficiency is achieved partly
because C. thermocellum assembles most of its
cellulase enyzmes onto an extracellular,
cell-associated scaffold. The entire assembly is
termed the cellulosome and maximizes synergies
between the different catalytic mechanisms of its
cellulase arsenal and subsequent sugar uptake.
Results and Discussion A constraint-based
genome-scale metabolic model has been constructed
based on the published annotation of Clostridium
thermocellums genome as well as the
incorporation of biochemical observation. The
statistics of the resulting model are shown in
the table to the right. The model is unique in
its incorporation of proteomics data to account
for substrate-dependent production of the
cellulosome.
Genome size 3.8 Mb
ORFs 3307
Included genes 432
Enzyme complexes 72
Isozyme cases 70
Reactions (excluding exchanges) 563
Transport 56
Gene associated 463
Non-gene associated intracellular 61
Non-gene associated transports 37
Distinct metabolites 529
Single gene deletions for which increased ethanol
production is predicted.
Comparison of model predictions to experimental
observations C. thermocellum iSR432 was used to
simulate growth in multiple conditions. Actual
(??) and predicted (?) reaction flux rates are
shown, and predicted fermentation product
production rates are shown as ranges as
determined by flux variability analysis. For
each simulation, the boundary fluxes for
cellobiose, acetate, and formate were constrained
to match the measured fluxes during (A) chemostat
growth on cellobiose and (B) fructose, and (C)
batch growth on cellobiose.
- References and Acknowledgements
- The authors would like to thank J. Paul Brooks
for his computational and operations expertise,
David Hogsett and Chris Herring for discussions
regarding C. thermocellum physiology, Lee Lynd
for generously providing C. thermocellum cultures
and Stephen Rogers and Evert Holwerda for
providing valuable assistance. - Lynd, L. R., Weimer, P. J., Van Zyl, W. H.,
Pretorius, I. S. (2002). Microbial cellulose
utilization fundamentals and biotechnology.
Microbiology and Molecular Biology Reviews,
66(3), 506-577. - Edwards, J. S., Covert, M., Palsson, B. Ø.
(2002). Metabolic modelling of microbes the
Flux-balance approach. Environmental
Microbiology, 4(3), 133-140. - Lamed, R. J., Lobos, J. H., Su, T. M. (1988).
Effects of Stirring and Hydrogen on Fermentation
Products of Clostridium thermocellum. Applied and
Environmental Microbiology, 54(5), 1216-1221. - Shlomi, T., Cabili, M. N., Herrgard, M. J.,
Palsson, B. Ø., Ruppin, E. (2008).
Network-Based prediction of human tissue-Specific
metabolism. Nature Biotechnology, 26(9),
1003-1010. - Burgard, A. P., Pharkya, P., Maranas, C. D.
(2003). Optknock a bilevel programming framework
for identifying gene knockout strategies for
microbial strain optimization. Biotechnology and
Bioengineering, 84(6), 647-657. - Fong, S. S., Burgard, A. P., Herring, C. D.,
Knight, E. M., Blattner, F. R., Maranas, C. D.,
et al. (2005). In silico design and adaptive
evolution ofescherichia coli for production of
lactic acid. Biotechnology and Bioengineering,
91(5), 643-648. - Desai, S., Guerinot, M., Lynd, L. (2004).
Cloning of l-Lactate dehydrogenase and
elimination of lactic acid production via gene
knockout in thermoanaerobacterium saccharolyticum
jw/sl-Ys485. Applied Microbiology and
Biotechnology, 65(5), 600-605.
The broad mixture of fermentation byproducts
produced by C. thermocellum is reflective of the
fact that it has evolved towards its own ends,
rather than for the production of ethanol. We
demonstrate here the creation of a genome-scale
metabolic model of the metabolism of C.
thermocellum and discuss its use for model-guided
metabolic engineering to optimize production of
ethanol from cellulosic substrates in C.
thermocellum.
Microbial Genomics 2009