E. Coli bacteria - PowerPoint PPT Presentation
1 / 49
Title:
E. Coli bacteria
Description:
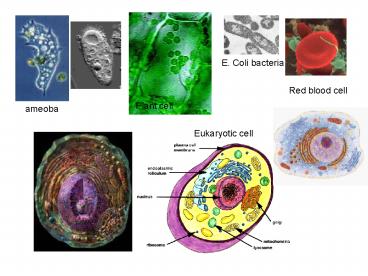
E. Coli bacteria Red blood cell Plant cell ameoba Eukaryotic cell HeLa cell with rhinovirus 4RHV.pdb 4RHV.pdb HEADER RHINOVIRUS COAT PROTEIN 25-JAN-88 ... – PowerPoint PPT presentation
Number of Views:293
Avg rating:3.0/5.0
Title: E. Coli bacteria
1
E. Coli bacteria
Red blood cell
Plant cell
ameoba
Eukaryotic cell
2
SEM images
3
(No Transcript)
4
(No Transcript)
5
(No Transcript)
6
_
_
Glycerol subunit
double bond
cholesterol
DPPC
1-palmitoyl-2-oleoyl-PC
1,2-di-palmitoyl-glycero-3 phosphocholine
160/181
160/160
7
(No Transcript)
8
Nanotech Application of DPPC (Coe Group)
shiny side Ni mesh
dull side Ni mesh
Thickness Ni mesh
10 mm
Cu-coated
10 mm
before Cu-coated
monolayer
trilayer
9
DPPC
metal
glycerol
chiral carbon
trilayer
monolayer
10
DPPC
chiral carbon
metal
11
Control of Optical Transmission
Through Microchannels
20o
30o
35o
40o
45o
50o
55o
60o
65o
70o
75o
20o
Optical Polarized Microscope Images of
Hexadecanethiol/Hydrated DPPC vs Temperature
Schematic of Coated Mesh
12
(No Transcript)
13
(No Transcript)
14
PM3 optimized Gramicidin A monomer (membrane
structure). 15 amino acids, alternating L and D,
coil into a nanochannel. A single-stranded,
helical dimer (HD) spans a membrane allowing ion
transport. O atoms (dark) of CO groups line the
ion channel.
Williams et al., Nanotechnology, 15 S495-S503
(2004)
15
CO stretch, 16 different stretches, Amide I bands
16
(No Transcript)
17
(No Transcript)
18
Halobacterium halobrium discovered by Dieter
Oesterhelt, Walther Stoeckenius and A. Blaurock
in 19711
Bacteriorhodopsin (BR) discovered by Dieter
Oesterhelt and Walther Stoeckenius in 19732
colonies framed by the main chromosome with the
external plasmids.
it is divided into one large chromosome with
2,014,239 bp and 2 small replicons pNRC100
(191,346 bp) and pNRC200 (365,425 bp).
O2 present they are red-pigmented4 as in a saline
pond at a salt work near San Quentin, Mexico
Halobacterium halobium sp. NRC-1 is a salt-loving
(Halophiles) archaebacterium that inhabits
natural salt lakes and areas where seawater is
evaporated to produce salt3.
BR in high concentrations but lacking O2 over San
Francisco Bay4,11 is purple, called purple
membranes
Red Halobacteria in Owens Salt Lake in Owens
Valley, California5
Halobacterium salinarum (electron microscope
image)9 0.5-1.2 um x 1.0-6.0 um in size10
Great Salt Lake in Utah, the Dead Sea, or Lake
Magadi in Southern Kenya's Maasai land
19
light
H
H
Purple membrane 2-D crystalline
bacteriorhodopsin lattice
ADP
flagellae
ATP
Sensor rhodopsins SR I and SR II
ATP-synthase
H
20
Archaea, common name for a group of one-celled
organisms, many of which do not require oxygen or
sunlight to live17.
Certain archaebacteria, members of a group of
primitive bacteria-like organisms, carry out
photosynthesis in a different manner. The
mud-dwelling green sulfur and purple sulfur
archaebacteria use hydrogen sulfide instead of
water in photosynthesis. These archaebacteria
release sulfur rather than oxygen, which, along
with hydrogen sulfide, imparts the rotten egg
smell to mudflats. Halobacteria, archaebacteria
found in the salt flats of deserts, rely on the
pigment bacteriorhodopsin instead of chlorophyll
for photosynthesis. These archaebacteria do not
carry out the complete process of photosynthesis
although they produce ATP in a process similar to
the light-dependent reaction and use it for
energy, they do not produce glucose. Halobacteria
are among the most ancient organisms, and may
have been the starting point for the evolution of
photosynthesis17.
21
The sequence of bacteriorhodopsin6,12
(CP)
The protein/lipid ratio is 7525
Aspartic Acid (Asp)
(EC)
Arginine (Arg)
Lysine (Lys)
22
(No Transcript)
23
(No Transcript)
24
Wt 26 kDa
Volume 83 nm3
5 nm
45 Å or 0.0045 mm
25
trans
9
11
7
5
13
4
8
10
15
12
6
3
14
2
1
Lysine (Lys)
hn 568 nm
9
7
11
5
13
4
8
cis
10
6
12
14
3
1
2
15
Lysine (Lys)
26
Photocycle of Bacteriorhodopsin6-7
BR568 hn
Photoisomerized to 13-cis
a 9-cis pathway19-20
J600
8-10 ms
500 fs
O640
Protonated all-trans
5 ps
K590
Retinal Quantum Efficiency in methanol 15.0
0.5 ms
2 ms
Retinal Quantum Efficiency in BR 67.0-64.0
N550
Solar Panels Efficiency 22.013
L550
2 ms
70 ms
M412 (CP)
Hin
M412 (EC)
Hout
De- and reprotonated of the Schiff base
27
Possible applications for long term recording,
3-D data and holographic storage19.
BR568 hn
Blue light to revert back
Q380
Thermal relaxation
8-10 ms
P490
O640
Photochemical activation
7
9
5
red pulse
4
10
8
6
9-cis pathway
11
12
3
1
2
14
13
15
28
Bacteriorhodopsin makes ATP by ATP-synthase.
It converts the energy of the light into an
electrochemical proton gradient (H ions
transferring across the membrane). The proton
gradient that results is used to drive ATP
synthesis by use of the ATP-synthase complex.
This modification allows bacteria to live in low
oxygen but rich light regions.
The H ions that are produced are then
transported outside of the cell. This results in
a potential energy gradient similar to that
produced by charging a flashlight battery. The
force the potential energy gradient produces is
called a proton motive force that can accomplish
a variety of cell tasks including converting ADP
into ATP8.
29
Proposed mechanism of light driven proton pumping
and conformational changes of bacteriorhodopsin7.
30
Applications of BR16
ID card with an optical data memory made from BR.
In the purple colored data strip, more than 1
MB of digital data may be stored permanently
A holographic camera for non-destructive Testing
using BR films as rewriteable Optical recording
media.
Prevent counterfeiting
ID-card as a novel security system
31
An important protein in the rod cell Rhodopsin15
Microscope images of Rod cells of a Zebrafish15
Wt 40 kDa
1 mm across
10 mm across
Cone cells important for color detection21
Red cones (560-566 nm max sensitivity)
Green cones (540-545 nm max sensitivity)
Blue cones (440 nm max sensitivity)
32
References and Sources used
1Oesterhelt, D. Stoeckenius, W. (1971) Nature
New Biol. 233, 149-152 and Blaurock, A.E.
Stoeckenius, W. (1971) Nature New Biol. 233,
152-155
2Dieter Oesterhelt and Walther Stoeckenius,
Functions of a new Photoreceptor Membrane Proc.
Nat. Acad. Sci. USA. 70, No 10, pp 2853-57, 1973
3http//biology.kenyon.edu/Microbial_Biorealm/arch
aea/halobacterium/halobacterium.html
4http//www.ucmp.berkeley.edu/archaea/archaealh.ht
ml
5http//waynesword.palomar.edu/plsept98.htm
6http//www.biochem.mpg.de/oesterhelt/
7Lubert Stryer Biochemistry 4th Edition, W.H.
Freeman and Company, New York, 1995
8http//www.creationresearch.org/crsq/articles/36/
36_1/atp.html
9http//www.biochem.mpg.de/oesterhelt/genomics/Int
ro_Hsal.html
10http//soils1.cses.vt.edu/ch/biol_4684/Microbes/
halo.html
11picture provide by Ruth Anderson
12http//www.ks.uiuc.edu/Research/Method/quant_sim
/
33
13http//www.qrg.northwestern.edu/projects/vss/doc
s/Power/2-how-efficient-are-solar-panels.html
14Feng Gai, K.C. Hasson, J. Cooper McDonald,
Philip A. Anfinrud. Chemical Dynamics in
Proteins The Photoisomerization of Retinal in
Bacteriorhodopsin, Science, Vol 27(20), March 20,
1998.
15http//www.accessexcellence.org/AE/AEC/CC/vision
_background.html
16http//www.chemie.uni-marburg.de/hampp/index_en
gl.htm
17http//beta.encarta.msn.com/encyclopedia_7615729
11_2/Photosynthesis.html
18http//www.tu-darmstadt.de/fb/ch/Fachgebiete/BC/
AKDencher/energie_en.html
19Norbert Hampp. Bacteriorhodopsin as a
Photochromic Retinal Protein for Optical
Memories, Chem Rev., Vol. 100, 1755-1776,
2000.
20Norbert Hampp, Nathan B. Gillespie, Kevin J.
Wise, Lei Ren, Jeffrey A. Stuart, Duane L.
Marcy, Jason Hillebrecht, Qun Li, Lavoisier
Ramos, Kevin Jordan, Sean Fyvie, and Robert R.
Birge. Characterization of the Branched-Photocycle
Intermediates P and Q of Bacteriorhodopsin,
J. Phys. Chem B., Vol. 106, 13352-13361, 2002.
21David W. Ball. The Baseline Eyes The Bodys
Own Spectroscopes, Spectroscopy, Vol. 20(4),
36-37, April 2005.
34
(No Transcript)
35
(No Transcript)
36
HeLa cell with rhinovirus
37
4RHV.pdb
38
4RHV.pdb
HEADER RHINOVIRUS COAT PROTEIN
25-JAN-88 4RHV 4RHV 3 COMPND
RHINOVIRUS 14 (/HRV14)
4RHV 4 SOURCE HUMAN (HOMO
SAPIENS) VIRUS GROWN IN HELA CELLS
4RHV 5 AUTHOR E.ARNOLD,M.G.ROSSMANN
4RHV 6 REVDAT
7 15-OCT-94 4RHVF 3 REMARK CRYST1 SCALE
4RHVF 1 REVDAT 6 15-JAN-92
4RHVE 1 REMARK
4RHVE 1 REVDAT 5 15-JUL-90 4RHVD 1
REMARK 4RHVD 1 REVDAT
4 15-JAN-90 4RHVC 1 REMARK
4RHVC 1 REVDAT 3 19-APR-89
4RHVB 1 SEQRES
4RHVB 1 REVDAT 2 09-OCT-88 4RHVA 1
JRNL 4RHVA 1 REVDAT
1 16-APR-88 4RHV 0
4RHV 7
39
(No Transcript)
40
(No Transcript)
41
(No Transcript)
42
(No Transcript)
43
(No Transcript)
44
(No Transcript)
45
(No Transcript)
46
1RHI.pdb
4RHV.pdb
Rhinovirus Coat Proteins, PDB files
47
(No Transcript)
48
DNA 101
- 3 Components of a Nucletide
- Nitrogenous base the major bases are
derivatives of 2 parent compounds - Pyrmidine 6 member ring (containing 2
nitrogens) - Adenine (A)
- Guanine (G)
- guanine
- Purine 6 member ring and 5 member ring
(containing 4 total nitrogens) - Thymine (T)
- Cytosine (C)
adenine
- 3 Components of a Nucletide
- Nitrogenous base the major bases are
derivatives of 2 parent compounds - Pyrmidine 6 member ring (containing 2
nitrogens) - Adenine (A)
- Guanine (G)
thymine
- cytosine
- 2-deoxy-D-ribose five carbon sugar residue with
carbons numbered using primes to distinguish them
from the carbons in the nitrogenous bases (1
carbon attaching to nucleic acid and 5 carbon to
phosphate bridge) - Phosphate bridge covalently links the 5
hydroxyl of one nucleotide to the 3 hydroxyl of
another
49
(No Transcript)