The experimental system PowerPoint PPT Presentation
Title: The experimental system
1
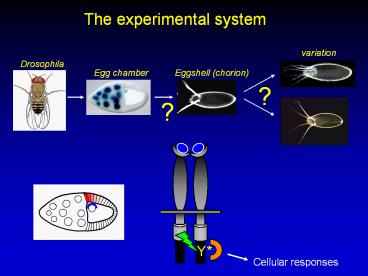
The experimental system
Cellular responses
2
EGFR/ErbB Signaling Network
Vertebrates
Invertebrates
ErbB3
EGFR
ErbB2
EGFR
signal diversification inhibitors ?
signal diversification heterodimerization
3
The EGFR network and dorsal patterning
Profile of EGFR activity WT
Initiation Gurken
4
How can we identify components of the EGFR
network?
5
Kek1 and feedback regulation of EGFR
EGFR
kek1 expression
?
No overt phenotype ?
6
Kek1 overexpression inhibits receptor signaling
ligand
Kek1
EGFR
7
Evidence that endogenous kek1 inhibits EGFR
signaling
8
What role does Kek1 have in dorsal patterning?
Initiation Gurken
Amplification Spitz Vein
Inhibition Argos
Site of Appendage Formation
9
Kek1 inhibits EGFR signaling
1. Mechanism 2. Sequence requirements
ligand
EGFR
Kek1
EGFR
10
Kek1 inhibits EGFR signaling
1. Mechanism
Ligand sink vs heterodimer
EGFR
11
Kek1 inhibits EGFR signaling
2. Sequence requirements ?
Domain swaps
12
Kek1 inhibits EGFR signaling
2. Sequence requirements ?
The Kek1 LRRs are sufficient for binding EGFR
but insufficient for full inhibition in vivo!
L
LIT
13
Identifying residues important for Kek1/EGFR
interaction
P
suppressor
F1
14
Identifying residues important for Kek1/EGFR
interaction
Knockouts of Kek1
15
Kek1 a novel inhibitory component of the EGFR
network
Binding Inhibition
EGFR
Kek1
Trafficking Stability
16
Identification of EGFR components
Genetic approach
Directed mosaics
Drosophila
Egg chamber
dCBP
bwk
WT
17
dCBP mediates feedback inhibition of EGFR
signaling
18
dCBP mediates feedback inhibition of EGFR
signaling
wt
Initiation Gurken
Amplification Spitz Vein
Inhibition Argos
Site of Appendage Formation
chorion
19
Egfr mediated epithelial patterning
Gurken
EGFR
Egfr transcriptional targets
20
Identification of EGFR components
Phylogenetic approach
4
latifasciaformis
21
EGFR signaling and appendage patterning
What components of the pathway underlie
variation in dorsal appendage patterning?
Intraspecific analysis
Interspecific analysis
map natural variation
Characterize patterning at molecular level
dose-dependent screen
? genes
22
Interspecific analysis Kek1
A
F
B
G
C
H
D
I
J
E
D. melanogaster
D. virilis
23
Intraspecific analysis
Does quantitative variation in the EGFR network
exist? If so, what molecular components
underlie this variation? - template for
interspecific variation?
Appendage variation
WT/V0 V1a V1b
V2 V3 V4
24
Intraspecific variation in EGFR signaling
25
EGFR signaling and appendage patterning
Identifying novel dose-dependent EGFR
components
grkHK36 egfrCO
26
Egfr mediated epithelial patterning
Gurken
EGFR
Egfr transcriptional targets
27
Acknowledgements
Andrea Cowden Omar Hasnie Matt Rubin Kyle Payne
Diego Alvarado Alex Doetsch Tim Evans Christina
MacLaren Amy Rice Anne Scherer
Fred Derheimer Libbie Eakin Paul
Miller Christopher Skipwith
Brandon Weasner