Generate mutants by mutagenesis of seeds
1 / 27
Title:
Generate mutants by mutagenesis of seeds
Description:
Map-based cloning of interesting genes. In a model organism ... 3. Narrow region down to a manageable length of DNA for cloning and sequence comparison ... –
Number of Views:130
Avg rating:3.0/5.0
Title: Generate mutants by mutagenesis of seeds
1
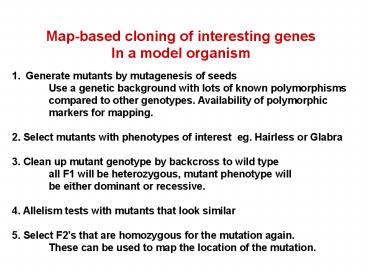
Map-based cloning of interesting genes In a model
organism
- Generate mutants by mutagenesis of seeds
- Use a genetic background with lots of known
polymorphisms - compared to other genotypes. Availability of
polymorphic - markers for mapping.
- 2. Select mutants with phenotypes of interest
eg. Hairless or Glabra - 3. Clean up mutant genotype by backcross to wild
type - all F1 will be heterozygous, mutant phenotype
will - be either dominant or recessive.
- 4. Allelism tests with mutants that look similar
- 5. Select F2s that are homozygous for the
mutation again. - These can be used to map the location of the
mutation.
2
GLABRA1 (GL1)
- Involved in trichome initiation
- Transcription factor
- Expressed in leaf primordia ? early trichome
initiation - gl1 mutants result in near complete loss of
trichome initiation
Wild type gl1 mutant
3
Scanning EM picture of Arabidopsis top leaf
surface with trichomes
4
Mapping Cross
Parents Col-0 gl/gl X La-er GL1/GL1 genotypes
F1 is self fertilized all chromosomes
recombine in meiosis F2 plants recombined
chromosomes segregate
How to do this with an organism that cannot
fertilize itself, like a mouse?
5
Mapping an Arabidopsis gene
- Analyze segregation data in an F2 population.
- Both chromosomes have had the opportunity to
become recombined in the F1 parent - To avoid confusion, we focus on one locus of
interest. - We chose individuals that are homozygous for one
allele at that locus, eg. a clear phenotype. - Closely linked markers will also be homozygous in
the chosen individuals. As markers are farther
away on the chromosome, more of the individuals
will have two different alleles for the marker
genes.
6
Interval mapping Identify markers linked to
the gene of interest that define an interval on a
chromosome.
7
Markers that define major regions of the
Arabidopsis chromosomes
8
F2s are selected as homozygous recessive gl1/gl1
by phenotype eg. Scored for 5 markers 1, 2 are
not linked to GL1 13 25 12 C/CC/LL/L Map
distance is calculated as recombinant
alleles/total X 100 cM 50 of alleles are C and
50 are L. Therefore the map distance from GL1
to 1 is 50 cm
9
Marker 1 from a previous year. The first lane is
the glabra mutant (Columbia), the second lane is
a mixture of DNA from lane 1 and lane 3 The
third lane is Landsberg The rest are DNA from F2
plants
10
We established that gl1 is on Chromosome 3.
What do we do next?
We can only find a locus by identification of
recombination events on either side. Identify
2 markers on Chromosome 3 that must be on
either side of gl1
11
Markers 3, 4 and 5 are linked to GL1 on
chromosome 3 We need to find another marker on
the opposite side of marker 3 to define the
interval that contains GL1.
3?
3?
gl1
gl1
3?
3?
4 or 5?
4 or 5?
12
3, 4, and 5 are linked to GL1 Marker 3 is
closest to GL1 Map distance is calculated as
recombinant alleles/total X 100 cM 3 is 4/100
X100 cM from GL1 4 cm 4 is 30 cM 5 is 20 cM
13
There is a recombination event between marker 3
and gl1 Which of the other 2 linked markers is
on the opposite side from marker 3? Plants 3,
4, 5 and 10 are useful to identify flanking
markers
14
Markers 3, and 5 flank GL1
Plant 3 is C/L at 3 L/L at 4 and C/C at 5
There has been no crossover between GL1 and
5 And 5 is further away from GL1 than 3. This
means 3 and 5 define the interval that contains
GL1.
Plant 3
5
gl1
3
Col-0
4
La-er
15
Marker 3 and 5 define an interval of 24 map
units that must contain gl1
16
Plants 3, 4 and 5 have recombination points
within the interval that defines the location of
GL1 They will be useful for further mapping
Plant 4
Plant 5
Plant 3 is C/L at 3 L/L at 4 and C/C at 5 Plant
4 is C/C at 3 C/C at 4 and C/Lat 5 Plant 5 is
C/L at 3 C/L at 4 and C/C at 5
Plant 3
Col-0
La-er
17
Identify more plants with recombination in the
interval
We will screen more F2 plants to identify those
with a recombination on either side of our
chosen interval to narrow in on the location of
the GL1 gene. We will analyze the alleles of
new markers located between marker 3 and marker
5. We will only analyze DNA from plants
heterozygous at either marker 3 or marker 5.
18
From comparison of genome sequence to a
recombination map made by Lister and Dean, we
learned that Arabidopsis has approximately 250
kb per map unit. That represents about 100
genes. For convenience, we aim for map
resolution of 0.1 map units, which should
represent 25-100 kb and hopefully 10-20 genes.
50-100 kb is the normal insert size for BAC
clones. In order to get to map resolution of
0.1 map units we screen at least 1000 F2 plants
(2000 chromosomes)
19
How to decide the number of F2s to examine?
Recombination frequency is calculated Number of
recombinants/number of chromosomes, 1
recombinant chromosome/2000 chromosomes 0.05
map units. We can only find a locus by
identification of recombination events on either
side. Therefore, with 2000 chromosomes we
should find one marker 0.05 cM to the right of
GL1 and another marker 0.05 cm to the left. An
interval of 0.1 map units between the two closest
markers is the best we measure. If we want
better resolution, we need more markers (which we
have) and more potential recombinant chromosomes
from F2 plants.
20
(No Transcript)
21
(No Transcript)
22
Once we have defined 2 markers flanking our
interval that are physically close enough, we
start sequence analysis for point mutations.
MDF20
MYN21
BAC T22A15 100 kb insert
BAC sequence gives us a list of genes. 20 in
Arabidopsis. GenBank annotation gives us a list
of predicted genes for each BAC from our ordered
library. Potential functions of the predicted
genes are defined by homology to other proteins.
Candidate genes can be chosen by predicted
function and expression pattern.
23
Expression pattern of genes in mapped interval
can help choose best candidate gene
24
Candidate genes can be PCR amplified from the
mutant and the sequence can be compared to wild
type. When a mutation is identified, we call
that a candidate gene. Transform mutant plant
with the wild type candidate gene for
complementation.
25
Alternatively, the entire BAC can be broken into
subclones. Each subclone can be used to
transform the mutant plant. If the BAC is made
with wild type DNA, subclones with the
correct gene in them will complement the
mutation.
26
Final confirmation
- Sequence mutant and wild type multiple mutant
alleles needed to be convincing - Complement mutation by making a transgenic with
the wild type copy of the candidate gene.
27
Finding a gene based on phenotype
- 1. 100s of DNA markers mapped onto each
chromosome high density linkage map. - 2. identify markers linked to trait of interest
by recombination analysis - 3. Narrow region down to a manageable length of
DNA for cloning and sequence comparison - 4. Compare mutant and wild type sequences to find
differences that could cause mutant phenotype - 5. Prove that mutation is responsible for
phenotype.