Restriction Enzymes - PowerPoint PPT Presentation
1 / 42
Title: Restriction Enzymes
1
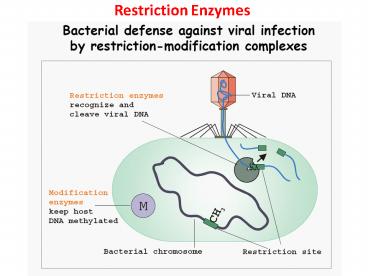
Restriction Enzymes
2
- Restriction Enzymes scan the DNA sequence
- Find a very specific set of nucleotides
- Make a specific cut
3
Palindromes in DNA sequences
- Genetic palindromes are similar to verbal
palindromes. A palindromic sequence in DNA is one
in which the 5 to 3 base pair sequence is
identical on both strands.
5
3
5
3
4
- Restriction enzymes recognize and make a cut
within specific palindromic sequences, known as
restriction sites, in the DNA. This is usually a
4- or 6 base pair sequence.
5
Restriction Endonuclease Types
- Type I- multi-subunit, both endonuclease and
methylase activities, cleave at random up to 1000
bp from recognition sequence - Type II- most single subunit, cleave DNA within
recognition sequence - Type III- multi-subunit, endonuclease and
methylase about 25 bp from recognition sequence
6
Hae III
- HaeIII is a restriction enzyme that searches the
DNA molecule until it finds this sequence of four
nitrogen bases.
5 TGACGGGTTCGAGGCCAG 3 3 ACTGCCCAAGGTCCGGTC 5
7
Once the recognition site is found Hae III will
cleave the DNA at that site
5 TGACGGGTTCGAGGCCAG 3 3 ACTGCCCAAGGTCCGGTC 5
8
These cuts produce blunt ends
5 TGACGGGTTCGAGG CCAG 3 3 ACTGCCCAAGGTCC GGTC
5
9
- The names for restriction enzymes come from
- the type of bacteria in which the enzyme is found
- the order in which the restriction enzyme was
identified and isolated.
- EcoRI for example
- R strain of E.coli bacteria
- I as it is was the first E. coli restriction
enzyme to be discovered.
10
blunt ends and sticky ends
- Hae III produced a blunt end?
- EcoRI makes a staggered cut and produces a
sticky end
5 GAATTC 3 3 CTTAAG 5
5 GAATTC 3 3 CTTAAG 5
5 G AATTC 3 3 CTTAA G 5
11
- blunt end
- sticky end
12
More examples of restriction sites of restriction
enzymes with their cut sites
- Hind III 5 AAGCTT 3
- 3 TTCGAA 5
- Bam HI 5 GGATCC 3
- 3 CCTAGG 5
- Alu I 5 AGCT 3
- 3 TCGA 5
13
Separating Restriction Fragments, I
14
Separating Restriction Fragments, II
15
Gene Cloning
- What is gene cloning? How does it differ from
cloning an entire organism? - Why is gene cloning done?
- How is gene cloning accomplished ?
- What is a DNA Library?
16
What is DNA cloning?
- When DNA is extracted from an organism, all its
genes are obtained - In gene (DNA) cloning a particular gene is copied
(cloned)
17
Why Clone DNA?
- A particular gene can be isolated and its
nucleotide sequence determined - Control sequences of DNA can be identified
analyzed - Protein/enzyme/RNA function can be investigated
- Mutations can be identified, e.g. gene defects
related to specific diseases - Organisms can be engineered for specific
purposes, e.g. insulin production, insect
resistance, etc.
18
How is DNA cloned?, I
Blood sample
- DNA is extracted- here from blood
- Restriction enzymes, e.g. EcoR I, Hind III, etc.,
cut the DNA into small pieces - Different DNA pieces cut with the same enzyme can
join, or recombine.
DNA
Restriction enzymes
19
The action of a restriction enzyme, EcoR I Note
EcoR I gives a sticky end
20
DNA Cloning, II
- Bacterial plasmids (small circular DNA additional
to a bacterias regular DNA) are cut with the
same restriction enzyme - A chunk of DNA can thus be inserted into the
plasmid DNA to form a recombinant
21
DNA cloning, III
- The recombinant plasmids are then mixed with
bacteria which have been treated to make them
competent, or capable of taking in the plasmids - This insertion is called transformation
22
DNA Cloning, IV
- The plasmids have naturally occurring genes for
antibiotic resistance - Bacteria containing plasmids with these genes
will grow on a medium containing the antibiotic-
the others die, so only transformed bacteria
survive
23
DNA Cloning, V
- The transformed bacterial cells form colonies on
the medium - Each cell in a given colony has the same plasmid
( the same DNA) - Cells in different colonies have different
plasmids ( different DNA fragments)
24
Screening, I
- Screening can involve
- Phenotypic screening- the protein encoded by the
gene changes the color of the colony - Using antibodies that recognize the protein
produced by a particular gene
25
Screening, II
- 3. Detecting the DNA sequence of a cloned gene
with a probe (DNA hybridization)
26
Polymerase Chain Reaction
- PCR
27
PCR
- invented by Karry B. Mullis (1983, Nobel Prize
1993) - patent sold by Cetus corp. to La Roche for 300
million - depends on thermo-resistant DNA polymerase (e.g.
Taq polymerase) and a thermal cycler
28
Heat-stable DNA polymerase
- Taq DNA polymerase was isolated from the
bacterium Thermus aquaticus. - Taq polymerase is stable at the high temperatures
(95oC) used for denaturing DNA.
Hot springs at Yellowstone National Park,
Wyoming.
29
DNA polymerase requirements
- template
- primer
- nucleotides
- regulated pH, salt concentration, cofactors
30
Steps in DNA replication
- template denatured
- primers anneal
- new strand elongation
31
Steps in a PCR cycle
- 1) template denatured
- 94 C, 30 sec
- 2) primers anneal
- 45-72 C, depending on primer sequence
- 30 sec 1 min
- 3) new strand elongation
- 72 C depending on the type of polymerase
- 1 min for 1000 nucleotides of amplified sequence
Number of specific DNA molecule copies grows
exponentially with each PCR cycle. Usually run
20-40 cycles to get enough DNA for most
applications (If you start with 2 molecules,
after 30 cycles you will have more than a billion)
32
PCR Process
- 25-30 cycles
- 2 minute cycles
- DNA thermal cycler
33
Template denatured
Annealing primers
New strand elongation
34
Uses for PCR
- Research
- Gene cloning
- Real-time PCR
- DNA sequencing
- Clinical
- DNA fingerprinting
- Crime scene analysis
- Paternity testing
- Archeological finds
- Genetically inherited diseases
35
DNA Sequencing
36
Chain termination method (Sanger Method),
sequence of single stranded DNA is determined by
enzymatic synthesis of complementary strands
which terminate at specific nucleotide
positions Chemical degradation method
(Maxam-Gilbert Method), sequence of a double
stranded DNA molecule is determined by chemical
treatment that cuts at specific nucleotide
positions
37
Dideoxynucleotide (ddNTP)
http//www.ncbi.nlm.nih.gov/books/bv.fcgi?ridhmg.
figgrp.604
38
http//www.ncbi.nlm.nih.gov/books/bv.fcgi?ridhmg.
figgrp.605
39
http//www.ncbi.nlm.nih.gov/books/bv.fcgi?ridhmg.
figgrp.605
40
http//www.ncbi.nlm.nih.gov/books/bv.fcgi?ridgeno
mes.figgrp.6477
41
http//www.ncbi.nlm.nih.gov/books/bv.fcgi?ridhmg.
figgrp.607
42
Costs and time for sequencing a human genome
(3.2 billion bp)