iProLINK: An integrated protein resource for literature mining and literaturebased curation
Title:
iProLINK: An integrated protein resource for literature mining and literaturebased curation
Description:
Access to iProLINK homepage. 4. iProLINK. http://pir.georgetown.edu/iprolink ... Extracted Annotations Tagged Abstracts ... Generation of domain expert-tagged corpora ... –
Number of Views:83
Avg rating:3.0/5.0
Title: iProLINK: An integrated protein resource for literature mining and literaturebased curation
1
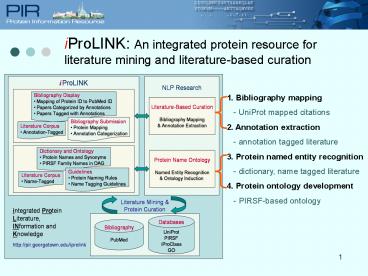
iProLINK An integrated protein resource for
literature mining and literature-based curation
1. Bibliography mapping - UniProt mapped
citations 2. Annotation extraction -
annotation tagged literature 3. Protein named
entity recognition - dictionary, name tagged
literature 4. Protein ontology development -
PIRSF-based ontology
2
Objective Accurate, Consistent, and Rich
Annotation of Protein Sequence and Function
- Literature-Based Curation Extract Reliable
Information from Literature - Function, domains/sites, developmental stages,
catalytic activity, binding and modified
residues, regulation, pathways, tissue
specificity, subcellular location ... - Ensure high quality, accurate and up-to-date
experimental data for each protein. - A major bottleneck!
- Ontologies/Controlled Vocabularies For
Information Integration and Knowledge Management - UniProtKB entries will be annotated using widely
accepted biological ontologies and other
controlled vocabularies, e.g. Gene Ontology (GO)
and EC nomenclature.
3
Access to iProLINK homepage
4
iProLINK
http//pir.georgetown.edu/iprolink/
- RLIMS-P text mining tool
- Protein dictionaries
- Name tagging guideline
- Protein ontology
5
Protein Phosphorylation Annotation Extraction
- Manual tagging assisted with computational
extraction - Training sets of positive and negative samples
Evidence attribution
RLIMS-P
3 objects
6
RLIMS-P Rule-based LIterature Mining System for
Protein Phosphorylation
download
http//pir.georgetown.edu/iprolink/
7
Benchmarking of RLIMS-P
High recall for paper retrieval and high
precision for information extraction
- UniProtKB site feature annotation
- Proteomics Mass Spec. data analysis protein
identification
8
Online RLIMS-P
(version 1.0)
http//pir.georgetown.edu/iprolink/rlimsp/
- Search interface
- Summary table with top hit of all sites
- All sites and tagged text evidence
9
BioThesaurus http//pir.georgetown.edu/iprolink/bi
othesaurus/
BioThesaurus v1.0
m million
(May, 2005)
10
BioThesaurus Report
Synonyms for Metalloproteinase inhibitor 3
- Gene/Protein Name Mapping
- Search Synonyms
- Resolve Name Ambiguity
- Underlying ID Mapping
1
3
ID Mapping
TMP3
Name ambiguity
2
11
Protein Name Tagging
- Tagging guideline versions 1.0 and 2.0
- Generation of domain expert-tagged corpora
- Inter-coder reliability upper bound of machine
tagging - Dictionary pre-tagging
- F-measure 0.412 (0.372 Precision, 0.462 Recall)
- Advantages helpful with standardization and
extent of tagging, reducing the fatigue problem,
and improve inter-coder reliability. - BioThesaurus for pre-tagging
12
PIRSF in DAG View
PIRSF-Based Protein Ontology
- PIRSF family hierarchy based on evolutionary
relationships - Standardized PIRSF family names as hierarchical
protein ontology - DAG Network structure for PIRSF family
classification system
13
PIRSF to GO Mapping
- Mapped 5363 curated PIRSF homeomorphic families
and subfamilies to the GO hierarchy - 68 of the PIRSF families and subfamilies map to
GO leaf nodes - 2329 PIRSFs have shared GO leaf nodes
- Complements GO PIRSF-based ontology can be used
to analyze GO branches and concepts and to
provide links between the GO sub-ontologies
14
Protein Ontology Can Complement GO
GO-centric view
- Expanding a Node Identification of GO subtrees
that can be expanded when GO concepts are too
broad - IGFBP subfamilies and
- High- vs. low-affinity binding for IGF between
IGFBP and IGFBPrP
15
Exploration of Gene and Protein Ontology
PIRSF-centric view
Molecular function
Biological process
- Systematic links between three GO sub-ontologies,
e.g., linking molecular function and biological
process - Estrogen receptor binding
- Estrogen receptor signaling pathway
16
Summary
- PIR iProLINK literature mining resource provides
annotated data sets for NLP research on
annotation extraction and protein ontology
development - RLIMS-P text-mining tool for protein
phosphorylation from PubMed literature. - BioThesaurus can be used for name mapping to
solve name synonym and ambiguity issues. - PIRSF-based protein ontology can complement other
biological ontologies such as GO.
17
Acknowledgements
- Research Projects
- NIH NHGRI/NIGMS/NLM/NIMH/NCRR/NIDCR (UniProt)
- NSF SEIII (Entity Tagging)
- NSF ITR (Ontology)
- Collaborators
- I. Mani from Georgetown University Department of
Linguistics on protein name recognition and
protein name ontology. - H. Liu from University of Maryland Department of
Information System on protein name recognition
and text mining. - Vijay K. Shanker from University of Delaware
Department of Computer and Information Science on
text mining of protein phosphorylation features.