Eukaryotic cell nucleus - PowerPoint PPT Presentation
1 / 31
Title:
Eukaryotic cell nucleus
Description:
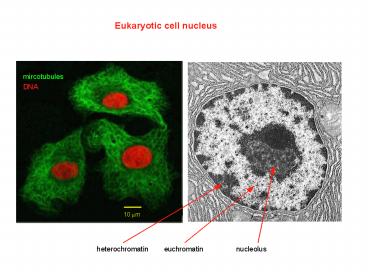
Eukaryotic cell nucleus mircotubules DNA 10 mm heterochromatin euchromatin nucleolus DNA Chromatin organization of higher eukaryotes Chromatin in the nucleus In 1973 ... – PowerPoint PPT presentation
Number of Views:512
Avg rating:3.0/5.0
Title: Eukaryotic cell nucleus
1
Eukaryotic cell nucleus
heterochromatin
euchromatin
nucleolus
2
(No Transcript)
3
Chromatin in the nucleus
- In 1884, Albrecht Kossel coined the term
histon to describe the proteins he - found by extracting avian erythrocyte nuclei
using diluted acids
- In 1973, Olins et al and Woodcock et al
observed that chromatin shows a - beads on a string structure by EM
- treatment of chromatin with micrococcal
nuclease preferentially cuts between the beads
4
Nucleosome structure
- Roger Kornberg
- based on EM images, nuclease digestion patterns,
X-ray diffraction data, and purification - of nucleoprotein complexes, proposed that the
nucleosome is the repeating unit of - chromatin and that every 200 bp of DNA forms a
complex with four histone pairs (1974)
5
Core histones
- core histones (H2A, H2B, H3 and H4) are small
(11 to 14 kD), highly basic proteins - they are evolutionarily highly conserved (from
yeast to humans) - they all share similar structural motifs
N
C
N-terminal tail
C-term tail
histone fold
hand shake motif
6
Assembly of a nucleosome
- histones can dimerize through their hand shake
motifs
- H3 can only dimerize with H4 and H2A always
dimerizes with H2B
- nucleosome assembly starts with two H3-H4
dimers forming a tetramer - this is followed by addition of two H2A-H2B
dimers to form the octamer - DNA is wrapped around the histone octamer
7
Nucleosome crystal structure
Luger et al, Nature, 1997
8
Nucleosome crystal structure
Luger et al, Nature, 1997
9
Why is chromatin folding important in the cell?
- DNA/chromatin has to condense and decondense
during the cell cycle
Stable cell line expressing H3-GFP
10
How does chromatin folding affect nuclear
functions?
- nucleosomes inherently function as barrier to
nuclear factors that need to - access and bind to DNA elements
- e.g. chromatinized template inhibits
transcription of underlying genes - also affects other DNA-templated processes such
as DNA replication, - repair etc.
- in order to activate gene expression, the cell
has developed ways to open - up chromatin
- ATP-dependent chromatin remodeling factors
- histone modifying enzymes
- insert histone variants at strategic locations
within genome
11
Post translational modifications on histones
- different modifications occur on specific
residues to perform specific - regulatory functions
12
Post translational modifications on histones
- Histone PTM has been a "hot research topic in
the last 15 yrs
Frequently asked questions
- What biological processes are associated
with/regulated by site-specific - histone modifications?
- What are the enzymes (acetylases, kinases,
methyl-transferases) that - directly modify histones at specific sites?
- What are the upstream pathways that regulated
these enzymes? - What are the downstream effects of histone PTMs
-- i.e. mechanism? - What are the enzymes that remove specific
histone PTMs? - What pathways that regulate these de-acetylases,
phosphatases, de-methylases etc?
13
Histone acetylation regulates transcription
activation
- It has long been known that histones in vivo
are acetylated, and as early as in - the 60s, Vincent Allfrey has suggested that
histone acetylation (and methylation) - regulate RNA synthesis
- e.g. by the 70s, Allfrey et al showed that
drugs that increase histone acetylation - in cells also increased DNase sensitivity of
the cellular DNA - by special labeling techniques, it was shown
that more accessible chromatin - are enriched for acetylated histones
- However, the direct link between histone
acetylation and transcription regulation wasnt - discovered till 1996 when the first
transcription-associated histone
acetyltransferase - (HAT) was identified
14
Identification of the first histone
acetyltransferase
The first transcription-associated histone
acetyltransferase (HAT) was identified by an in
gel histone acetyltransferase assay
histone substrates
SDS PAGE
cut out for peptide sequencing, protein ID
denature and renature proteins in the gel
3H Ac-CoA
Coomassie stain
Autorad
Brownell et al, Cell, 1996
15
Transcription is regulated by the balance of HATs
and HDACs
- The first HAT identified was Gcn5, which was a
well-studied transcription - co-activator identified by genetics studies in
yeast - Also in 1996, the first histone deacetylase
(HDAC) was identified, and the enzyme - Rpd3 was also a long studied transcription
repressor identified by yeast genetic - studies
- Many other transcription co-activators and
repressors were found to be HATs and - HDACs respectively, and these enzymes are
recruited to promoters during - transcription activation or repression
16
Technical advances that helped the study of
histone modifications
- 1. Development and refinement of in vitro
assays
radioactive co-factor
enzyme source
substrate
modified histones
3H-Ac-CoA (acetylation)
nuclear extracts
histones
3H-SAM (methylation)
IPd protein
nucleosomes
32P-ATP (phosphorylation)
recombinant protein
peptides
32P-NAD (ADP-ribosylation)
17
Example identification of a histone H3
methyltransferase
fractionate nuclear lysates by chromatography
techniques
collect fractions
add histone H3 substrate and 3H SAM
separate proteins by SDS PAGE stain gel or do
autoradiography identify fractions that contain
radio-actively labeled H3 repeat fractionation if
necessary identify histone modifying enzyme
Wang et al, Mol Cell, 2001
18
How to identify site of histone modification?
19
How to identify site of histone modification?
histone methyl- transferase
H3 peptide
3H-SAM
radioactively-labeled peptide
protein sequencing (Edman degradation)
detect radioactive amino acid
Strahl et al, PNAS, 1999
20
Technical advances that helped the study of
histone modifications
- Development and usage of histone
modification-specific antibodies
- antibodies are very useful reagents for
research - they can have exquisite specificities and
sensitivities for detection of proteins - can generate and purify antibodies that
specifically detect site-specifically - modified histones
21
Technical advances that helped the study of
histone modifications
- Development and usage of histone
modification-specific antibodies
22
Recent article in BMC Bioinformatics on
epigenetics and histone modifications
23
Technical advances that helped the study of
histone modifications
- Development and usage of histone
modification-specific antibodies
- antibodies are very useful reagents for
research - they can have exquisite specificities and
sensitivities for detection of proteins - can generate and purify antibodies that
specifically detect site-specifically - modified histones
- these antibodies can be used for Western blot
analyses, immunofluorescence - (IF) studies, and chromatin immunoprecipitation
(ChIP) assays
24
Uses of modification-specific histone antibodies
- 2a. Western blot analyses
- modification-specific histone antibodies are
useful for monitoring - overall abundance and global changes of
specific histone modifications
Briggs et al, Genes Dev, 2001
25
Uses of modification-specific histone antibodies
- 2b. Immunofluorescence assays
- modification-specific histone antibodies can be
used to examine localization - of the modified histones within the nucleus
Chromosome enriched in Lys9-methylated H3
Me(Lys9) H3
26
Uses of modification-specific histone antibodies
- 2c. Chromatin immunoprecipitation assay
- ChIP assay is useful for examining the
enrichment of specific histone- - modifications or binding of specific factors
to the gene of interest in vivo
27
Uses of modification-specific histone antibodies
- 2c. Chromatin immunoprecipitation (ChIP) assay
- can be coupled to gene activation procedures to
look at changes in histone- - modifications or transcription factor binding
to specific genes before and after - transcription activation
- can also be used in combination with microarray
analyses (ChIP on chip) or deep- - DNA sequencing (ChIP-seq) to do genome-wide
mapping of histone modifications - and chromatin-binding proteins
- while ChIP-chip or ChIP-seq provide
correlational information, detailed ChIP - analyses of specific genes can help eludicate
step-wise mechanisms
28
Transcription activation of the b-interferon gene
- The b-interferon gene is highly activated upon
viral infections and has served as a - model system to study gene activations
mRNA levels
ChIP assays
adapted from Agalioti et al, Cell, 2000
29
How does histone acetylation promote
transcription?
- Acetylation neutralizes the positively charged
lysine residues on histones and thus - reduces the interactions of the histones with
the negatively charged DNA
- Acetylated histones recruit and stabilize
binding of transcription or chromatin - remodeling factors via interactions of the
acetylated lysines with the - Bromodomains of these nuclear factors
Jacobson et al Science 2000
30
(No Transcript)
31
Different dynamics of histone modifications
highly dynamic
HMT
more stable
histone
Me-histone
de-methylase