Forward Genetics - PowerPoint PPT Presentation
1 / 19
Title:
Forward Genetics
Description:
Forward Genetics Begins with a mutant (altered) phenotype and addresses the question, what is the genotype? or mutation in which gene caused the alteration in ... – PowerPoint PPT presentation
Number of Views:350
Avg rating:3.0/5.0
Title: Forward Genetics
1
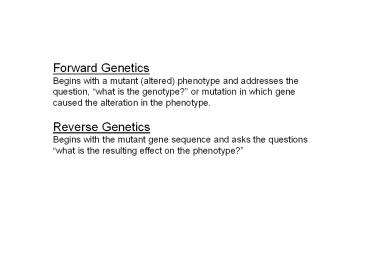
Forward Genetics Begins with a mutant (altered)
phenotype and addresses the question, what is
the genotype? or mutation in which gene caused
the alteration in the phenotype. Reverse
Genetics Begins with the mutant gene sequence and
asks the questions what is the resulting effect
on the phenotype?
2
Genome sequencing has provided means to carry out
systematic reverse genetics in large scale. Gene
knock-out or null mutation provide direct means
of studying the function of a gene product in
situ.
Arabidopsis is 120 Mb, contains small introns and
small intergenic regions, therefore there is a
high chance of finding mutation in every gene in
T-DNA insertion population.
3
"Knockology."
4
SATURATING THE GENOME WITH MUTATIONS
Three variables determine the probability that a
T-DNA insert will be found within a given gene
the size of the gene, the size of the genome, and
the number of T-DNA inserts distributed among the
population
Curves are drawn for several different gene
lengths 0.5, 1, 2, 3, 4, and 5 kb. The
probabilities were calculated using the following
formula P 1 - (1 - x/120,000)n, where P
probability of finding one T-DNA insert within a
given gene, x length of the gene in kilobases,
and n number of T-DNA inserts present in the
population. This calculation assumes that the
haploid Arabidopsis genome is 120 Mb and that
T-DNA insertion is random.
5
Organization and Screening of 60,480
T-DNATransformed Arabidopsis Lines. (A) Pooling
strategy. (B) Insertion screening strategy. 5'
and 3' refer to PCR primers specific for the gene
of interest. T-DNA L and T-DNA R refer to PCR
primers specific for the T-DNA border regions.
kanr, kanamycin resistant.
6
Alternatives to insertion mutations
Why? First, many genes are functionally
redundant, sharing overlapping functions with
other genes that may or may not be related at the
sequence level. Mutation of a functionally
redundant gene is not likely to lead to an easily
recognizable phenotype, because one or more other
family members can provide the same function.
Analysis of systematic gene knockouts has
revealed that a significant percentage of yeast
genes have no obvious phenotype when disrupted,
despite testing under a wide range of growth
conditions. Therefore, it is likely that
disruption of many plant genes will not result in
an easily identifiable phenotype. Second, many
genes function at multiple stages of development.
Mutations in these genes may lead to early
lethality or may be highly pleiotropic, which can
mask the role of a gene in a specific pathway.
7
GUS Reporter Gene Expression Patterns in
Representative Enhancer or Gene Trap Lines.
Expression is evident, as follows (A) In
cotyledons and shoot apex but not leaves. (B) In
trichomes. (C) In stipules and leaf tips. (D)
In a single cell at the tip of leaf primordium.
(E) In a lateral root primoridium. (F) In a
root tip. (G) In a root cap.
8
Gene traps allow the identification of genes that
are not amenable to classical genetic analysis.
Therefore, novel genes are likely to be found in
any gene trap screen. Screens have in fact been
successful in identifying genes specifically
expressed in lateral roots, developing embryos,
and shoot apices. Conditional screens have also
been performed to identify genes regulated by
nematode infections and in response to abiotic
stress.
The PROLIFERA (PRL) gene was identified as a gene
trap DsG insertion that showed GUS activity in
dividing cells. PRL encodes a protein that is
related to MCM7, a member of the MCM gene family
found in all eukaryotes and required for the
initiation of DNA replication. Expression in
dividing cells is consistent with this predicted
role in cell division. Disruption of PRL by the
DsG element led to megagametophyte and embryo
lethality. Arrest of both megagametophytes and
embryos occurred at variable stages of
development. There are many embryo-lethal
mutations in Arabidopsis that have variable
phenotypes, which makes it difficult to determine
the cause of lethality. However, the GUS
expression pattern in dividing cells suggested a
role in cell division before the gene was cloned.
9
Establishing an efficient Ac/Ds tagging system in
rice large-scale analysis of Ds flanking
sequences
Ac and Ds T-DNA vectors. RB and LB, right and
left borders of T-DNA 35S, CaMV 35S promoter
Ac, immobilized transposon Ubi, maize ubiquitin
promoter gfp, green fluorescent protein HPT,
hygromycin phosphotransferase gusA,
-glucuronidase gene bar, phosphinothricin
acetyltransferase gene Act1, rice actin
promoter I, intron from Arabidopsis G protein
gene A, triple splice acceptor nos3' and ocs3',
nopaline and octopine 3' UTR (poly(A) signal).
10
Large-scale generation of stable unlinked Ds
transposants. Ds element was mobilized by
crossing homozygous Ds and Ac lines F1 seeds
were planted and allowed to self-pollinate. In F1
generation, Ds transposed to a new locus in the
genome, germinated F2 seeds were screened against
GFP-linked donor sites, and the GFP seeds
were planted on soil. At the three-leaf stage,
plants were sprayed with Basta to select
transposants. DNA was isolated from F2 plants,
and bar-PCR was carried out to confirm the
presence of Ds element.
11
Distribution of genetically mapped Ds sites
The results indicated that different Ds elements
showed preference for certain chromosomes Ds3
(from chromosome 2) had a statistically
significant preference for chromosome 7 Ds4
(from chromosome 1) for chromosome 6 Ds5 (from
chromosome 8) and Ds9 (from chromosome 10) for
chromosome 1
12
PTGS for functional genomics
RNAi Injection of dsRNA into nematodes can
trigger specific (homologous) RNA degradation.
RNAi has been utilized to study 4000 genes in C.
elegans. In plants, dsRNA and sshairpin RNA have
similar effect.
The predicted RNA structure and efficacy of
gene-silencing constructs.
(Ref Wesley et al., 2001, Plant J. 27 581-590)
Constructs, controlled by Ubi1 promoter,
silencing GUS in rice. Thick lines indicate a 560
nt GUS sequence grey lines indicate non-GUS
sequences dashed grey lines indicate
intron-junction sequences left after splicing
and short lines within the stem of hairpin
structures indicate base pairing. Numbers in PTGS
column indicate the percentage of plants showing
GUS silencing n number of plants in each
treatment.
13
From Ref Wesley et al.
(Ref Wesley et al., 2001, Plant J. 27 581-590)
Using hpRNA constructs, we have obtained silenced
plants for every gene that we targeted,
irrespective of whether it was a viral gene,
transgene or endogenous gene, and the silencing
appears to be uniform within tissues in which
the hpRNA is expressed. With ihpRNA constructs
the efficiency averaged about 90, and arms of
400800 nt appear to be stable and effective.
High levels of silencing were obtained with
constructs having unmatched arm lengths, with
arms as long as 853 nt or as little as 98 nt, and
with arm sequences derived from coding, 3 or 5
untranslated regions of the target gene. These
results suggest that ihpRNA constructs will be
effective in a wide range of circumstances, and
augur well for the generic use of the technology.
The silencing was much more profound with ihpRNA
constructs than either anti-sense
or co-suppression constructs some ihpRNA
transformants were close to exhibiting a
complete knockout of the target endogenous gene.
However, most of the ihpRNA plants showed
dramatically reduced but detectable levels of
target gene activity.
14
VECTORS FOR VIRUS-INDUCED GENE SILENCING The
regions of the plant that can be affected by gene
silencing depend mostly upon viral vector
characteristics. Silencing vectors usually travel
systemically through the phloem in the vascular
tissues to most parts of the plant. Although most
of the viruses do not penetrate meristems, some
have been found to deliver a silencing signal to
meristematic regions of the plant. Some important
characteristics of a viral silencing vector are
its effectiveness in inducing silencing its
capability to infect and induce silencing in
growing parts of the plant its genome size (due
to cloning steps for fragment insertion, since
usually the smaller the vector the easier the
cloning) its genome partition the type of
nucleic acid its genome is composed of (given
that RNA plant viruses are the most common,
though DNA viruses can facilitate inoculation as
they allow the use of DNA instead of unstable
RNA) its host range whether its genome is or
can be made available in binary vectors (to
facilitate inoculation via Agrobacterium), and
its safety for individuals and the environment
15
Tobacco mosaic virus (TMV) was the first viral
vector used to successfully elicit VIGS of an
endogenous gene in a plant species. Potato
virus X (PVX) followed as the next viral vector
used to carry a gene into a plant cell and
trigger silencing via VIGS. Although effective,
its incapability of infecting meristems is a
great disadvantage, since delivering silencing to
meristematic tissues is a sine qua non for
studying genetic functions involved in
developmental pathways. The advent of the
Tobacco rattle virus (TRV) vector for VIGS opened
the possibility of studying genes expressed in
early organ development, because of its ability
to reach growing points and to deliver the
silencing signal to meristems. The cloning of its
genome into binary Agrobacterium tumefaciens
plasmids immensely facilitated the infection
process
16
TRV is an ssRNA virus with a bipartite genome.
The component called RNA1 encodes, among other
genes, an RdRP, whereas its genome partner,
called RNA2, encodes the coat protein. The target
gene fragment for silencing is inserted into the
RNA2 element. Inoculation, either mechanical or
via agroinfiltration, requires the presence of
both genome components. In the case of
agroinfiltration, two different Agrobacterium
clones, one carrying the RNA1 genome and another
with the RNA2 containing the target gene
fragment, are mixed together and co-infiltrated
into the leaf tissues.
17
A significant limitation of the VIGS system is
that TRV, and most of the other vectors
available, work very well in N. benthamiana and
in the tomato, but they do not show efficacy in
Arabidopsis, in which a myriad of genes still
need to be functionally analyzed. Additionally, a
vector with the capability to infect monocot
species, such as maize, rice and lily, is also
greatly desired.
18
Silencing of two endogenous genes was achieved
from DNA fragments carried in different TGMV
component vectors. Variegation occurred in
leaves that were partly expanded at the time of
inoculation, however, very little stem elongation
was evident in new growth (k). Plant (l)
inoculated with A790su/B122PCNA and pruned
(arrow) 2 weeks after inoculation showed
silencing in axillary buds. PCNA silencing is
evidenced by reduced stem elongation and aberrant
leaf formation. The two axillary outgrowths show
different degrees of su silencing with one
cluster of leaves (right) showing almost no
chlorophyll. Note circular yellow spots in
inoculated lower leaves (black arrow).
Silencing of a meristematic gene using
geminivirus-derived vectors
19
Silencing of the PDS gene. Infection of
recombinant TRV carrying the PDS (phytoene
desaturase) sequence silences endogenous PDS in
N. benthamiana plants and causes inhibition of
carotenoid biosynthesis resulting in the
photobleaching phenotype. On the left, whole
plant and on the right, enlarged single leaf.