SOURCE ORIENTED vs RECEPTOR ORIENTED MODELS - PowerPoint PPT Presentation
1 / 55
Title:
SOURCE ORIENTED vs RECEPTOR ORIENTED MODELS
Description:
... for fitting species divided by the degrees of freedom yields the chi square. The highest R/U values for fitting species are the cause of high chi square values. ... – PowerPoint PPT presentation
Number of Views:276
Avg rating:3.0/5.0
Title: SOURCE ORIENTED vs RECEPTOR ORIENTED MODELS
1
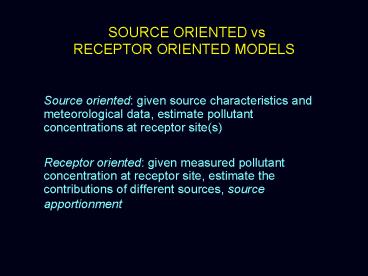
SOURCE ORIENTED vs RECEPTOR ORIENTED MODELS
- Source oriented given source characteristics and
meteorological data, estimate pollutant
concentrations at receptor site(s) - Receptor oriented given measured pollutant
concentration at receptor site, estimate the
contributions of different sources, source
apportionment
2
Receptor models
- CMB Chemical Mass Balance
- Factor Analysis (Multivariate methods)
- PCA Principal Component Analysis
- PMF Positive Matrix Factorization
3
Chemical Mass Balance Receptor Modelling
Source 1xi1 i1,n
3
Receptoryi i1,n
1
Source 3xi3 i1,n
2
Source 2xi2 i1,n
4
Components
- PM
- metals
- ions
- PAH
- OC/EC
- etc.
- Gas phase
- various organic cpds
- VOCs
- etc.
5
Chemical Mass Balance Receptor Modelling
6
Chemical Mass Balance Receptor Modelling
7
Chemical Mass Balance Receptor Modelling Simple
Spreadsheet Implementation
- Electronic spreadsheets like Excel make it easy
to implement the model in the above form. - The Solver function in Excel enables the
minimization operation. - Graphical tools in Excel enable qualitative
assessment of model fit and the inputs.
8
(No Transcript)
9
(No Transcript)
10
(No Transcript)
11
(No Transcript)
12
U.S. EPAs CMB8
- The CMB model incorporates the statistics to
improve our estimate of the source contributions
by weighting the source and receptor profiles
according to the uncertainties associated with
them. - It also reports the uncertainties associated with
the source contribution estimates and goodness
of fit parameters. - Version 8 is windows based, more user friendly.
- Example Source apportionment for motor vehicle
related VOCs in micro-environments
13
Chemical Mass Balance Receptor Modelling U.S.
EPAs CMB Model
- There are uncertainties associated with both the
receptor and source profiles. The measured
quantities are thus - The model results then also have uncertainties
associated with them
14
U.S. EPAs CMB Model The effective variance
method
- The CMB model uses an effective variance which
looks at variances in both the receptor and
source profiles, the latter weighted by the
source contributions. The inverse of these are
then used to weight the discrepancies for each
element, I.e. the contribution of a component
with smaller effective variance will be weighted
more in the iterative procedure.
15
CMB Effective Variance Iterative
procedure(simplified)
- Start with all source contributions 0
- Find the elements of the effective variance
matrix - Estimate new source contribution estimates that
will reduce the sum of the square of the errors
weighted by the elements of the effective
variance matrix. - Check the new source estimates against previous
ones - Go to 5 if all are within 1 of old values
- Otherwise replace them with new values and go
to 2. - 5. Assign the last source contribution
estimates, calculate statistical performance
indicators and report
16
Sources
17
Species
18
(No Transcript)
19
(No Transcript)
20
(No Transcript)
21
Fitting sources and species
- The receptor data may contain concentrations of
many species, some of which may not be
particularly interesting for our source
apportionment. - We may also have data on many source types, only
some of which have practical significance for a
particular receptor. - We may therefore choose the relevant species and
sources as fitting, I.e. those that will be
included in the iterative refinement procedure.
The remaining sources will not be considered at
all. The mass balances on the remaining species
will be calculated and reported but will not
affect the source apportionment.
22
Receptors
- The last two columns are x-y coordinates of
receptors for graphical display of results
23
Source contribution estimates SCE
24
STD ERR
- The standard errors reflect the precision of the
ambient data, the source profiles, and the amount
of collinearity among different profiles. - There is about a 66 probability that the true
source contribution is within one standard error
and about a 95 probability that the true
contribution is within two standard errors of the
source contribution estimate.
25
TSTAT
- The T-statistic (TSTAT) is the ratio of the
source contribution estimate to the standard
error. - A TSTAT value less than 2.0 indicates that the
source contribution estimate is at or below a
detection limit.
26
Performance measures
- R SQUARE
- PERCENT MASS
- CHI SQUARE
- DF
27
CHI SQUARE
- The chi-square is the weighted sum of squares of
the differences between the calculated and
measured fitting species concentrations. - The weighting is inversely proportional to the
squares of the precision in the source profiles
and ambient data for each species. Ideally, there
would be no difference between calculated and
measured species concentrations and chi-square
would equal zero. - A value less than 1 indicates a very good fit to
the data, while values between 1 and 2 are
acceptable. Chi-square values greater than 4
indicate that one or more species concentrations
are not well explained by the source contribution
estimates.
28
DF
- The degrees of freedom equal the number of
fitting species minus the number of fitting
sources. The degrees of freedom is needed when
statistical significance tests are applied to the
chi-square value.
29
R SQUARE
- The R-square is the fraction of the variance in
the measured concentrations that is explained by
the variance in the calculated species
concentrations. - It is determined by a linear regression of
measured versus model-calculated values for the
fitting species. - R-square ranges from 0 to 1.0. The closer the
value is to 1.0, the better the source
contribution estimates explain the measured
concentrations. - When R-square is less than 0.8, the source
contribution estimates do not explain the
observations very well with the fitting source
profiles and/or species.
30
PERCENT MASS
- Percent mass is the percent ratio of the sum of
the model-calculated source contribution
estimates to the measured mass concentration.
This ratio should equal 100, although values
ranging from 80 to 120 are acceptable. If the
measured mass is very low (lt 5 to 10 ?g/m3),
percent mass may be outside of this range because
the precision of the mass measurement is on the
order of 1 to 2 ?g/m3.
31
Combined Performance Measure
- The Fit Measure is calculated using the
algorithm - Fit_measure Wt_chisqr ( 2 / chisqr )
- Wt_R-square R-square
- Wt_pcmass pcmass / 100 (for pcmass lt 100) or
- Wt_pcmass 100 / pcmass ( for pcmass gt 100)
- Wt_fracest Frac_Est
- where chisqr, rsquare, and pcmass are the
performance measures for reduced chi-square, R-
square, and per cent mass, respectively, and
where Frac_Est is the ratio of the number of
estimable fitting sources to the total number of
fitting sources. - The weights accorded to each of these variables
(Wt_chisqr, Wt_R-square, Wt_pcmass , and
Wt_fracest) are set to 1.0 by default. These
weights may be changed in the Options window.
32
Source contributions to species
- Recall that the basis of the CMB model is that
species are transported equally effectively from
the source to the receptor (i.e. conservative
species) - However, because each component is present in
different relative abundances in the various
sources, the contribution of different sources to
the receptor load for a particular component
will be different for each component. - Furthermore, the mass balance closure for each
component will be different.
33
(No Transcript)
34
- This display shows how well the individual
ambient concentrations are reproduced by the
source contribution estimates. It offers clues
concerning which sources might be missing or
which ones do not belong in the calculation. - Fitting species are marked with an asterisk in
the column labeled ' I '. - Note that the symbol lt flags values less than the
uncertainty. - The asterisks indicate numerical overflow,
meaning that the number is too large to be
displayed in the allocated field.
35
- The column labeled RATIO R/U contains the ratio
of the signed difference between the calculated
and measured concentrations (the residual)
divided by the uncertainty of that residual
(square root of the sum of the squares of the
uncertainty in the calculated and measured
concentrations). - The R/U ratio specifies the number of uncertainty
intervals by which the calculated and measured
concentrations differ. - When the absolute value of the R/U ratio exceeds
2, the residual is significant. If it is
positive, then one or more of the profiles is
contributing too much to that species. If it is
negative, then there is an insufficient
contribution to that species and a source may be
missing. - The sum of the squared R/U for fitting species
divided by the degrees of freedom yields the chi
square. The highest R/U values for fitting
species are the cause of high chi square values.
36
(No Transcript)
37
MPIN
- The Modified Psuedo Inverse (MPIN) matrix shows
which species have the largest influence on the
source contribution estimates from each profile. - MPIN is normalized such that it takes on values
from -1 to 1. Species with MPIN absolute values
of 1 to 0.5 are considered influential species.
Noninfluential species have MPIN absolute values
of 0.3 or less. - Species with absolute values between 0.3 and 0.5
are ambiguous but should generally be considered
noninfluential.
38
MPIN matrix
39
VOC's Associated with Motor Vehicles
Exhaust EmissionsEvaporative Emissions
C1 - C10 RangeAlkanesAlkenesAlkynesCycloalkane
sAromatics
40
Chemical Mass Balance Receptor Modelling
Source profile characterization
Fuel
Whole gasoline
Headspace vapor
Individual vehicles
Exhaust emissions - UDDS
Evaporative emissions - SHED
Fleet emission characteristics
Tunnel studies Roadside measurements
41
Source Profiles
42
ERMD Source Profiles with 17 species
0.45
0.4
0.35
0.3
0.25
Fraction of total
0.2
0.15
0.1
6 GA
0.05
5 LPG
4 CNG
0
3 HOTST
2 COLDST
1 FUEL
Ethylene
Ethane
Propane
n-butane
n-pentane
benzene
3m-pentane
e-benzene
o-xylene
43
1994 Fleet Average Emissions with two fuels
44
Ozone forming potential of exhaust from 1994
Fleet
45
Modelled Species Profile
3m-pentane m-cyclopentanebenzene
cyclohexane iso-octanen-heptane toluene
n-octane e-benzenemp-xylenen-nonane1,2,4-t
m-benzene
ethylene acetylene ethane propane
isobutane isobutylenen-butane 2m-butane
n-pentane 2,3-dm-butane 2m-pentane
Fitting species
46
Nose-level Ambient Sampling Schedules
Slater Street Curbside 20 Weekdays in
August-September
2 Stations, 3 Sampling Sessions Morning
730-930Noon 1130-1330Afternoon
1530-1730 (Evening 2200 - 2400)
- 3 Underground Garages
- 12 Weekdays in December-January
- 2 Stations, 2 Sampling sessions in each garage
- A.M. 730 - 930
- P.M. 1500 - 1700
47
(No Transcript)
48
(No Transcript)
49
(No Transcript)
50
- Watson, J.D., E. Fujita, J.C. Chow, and B.
Zielinska. 1998. Northern Front Range Air Quality
Study Final Report., Desert Research Institute.
51
Other receptor models
- CMB applies the mass balance principles to known
receptor and source compositions to arrive at the
source contribution estimates. - Another type of source apportionment analyzes
multiple measurements at the receptor (I.e. many
hours of hourly average data) without prior
knowledge of the sources but attempts to identify
factors that explain the variation in the
composition. The factors are then interpreted as
types of sources.
52
Positive Matrix Factorization, PMF
53
Positive Matrix Factorization, PMF
- The rows of the F matrix then are interpreted to
be the profiles of factors (sources), and the
rows of the G matrix are the contributions of
each source for that observation period.
54
CMB PMF Comparison
- Even a single receptor observation can be
analyzed by CMB, assuming we have characterized
the potential sources. - PMF does not require that we characterize the
potential sources but it does require sufficient
observations to identify the potential sources.
55
The SPECIATE database
- Source profiles for PM and VOC samples are
compiled in a database (SPECIATE) which reports
measurements from different studiES. - The profiles in this database can be extracted
for use with CMB.