Many human genetic diseases are caused by mutations causing missplicing
1 / 54
Title:
Many human genetic diseases are caused by mutations causing missplicing
Description:
Alternative 5' splicing. Unwanted = longer exon. Antisense shorter isoform ... B. Bias alternative splicing ratios. Target the unwanted isoform exon-intron joint. ... –
Number of Views:339
Avg rating:3.0/5.0
Title: Many human genetic diseases are caused by mutations causing missplicing
1
Many human genetic diseases are caused by
mutations causing missplicing
Last updated 12/9/06 1 PM
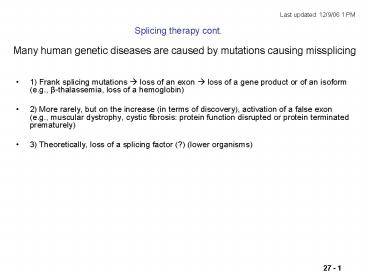
Splicing therapy cont.
- 1) Frank splicing mutations ? loss of an exon ?
loss of a gene product or of an isoform (e.g.,
ß-thalassemia, loss of a hemoglobin) - 2) More rarely, but on the increase (in terms of
discovery), activation of a false exon (e.g.,
muscular dystrophy, cystic fibrosis protein
function disrupted or protein terminated
prematurely) - 3) Theoretically, loss of a splicing factor (?)
(lower organisms)
2
Therapeutic intervention at the level of pre-mRNA
splicing
Alternative splicing Unwanted alternative
included Use antisense ? skipped Bias alternative
splicing Against an unwanted isoform (e.g.,
Bcl-X alt. spl. Bcl-XS promotes
apoptosis Bcl-XL inhibits apoptosis and
promotes cell growth, cancer)
Pseudo exon activated ? disease Antisense block
and skip unwanted pseudo exon
Alternative 5 splicing Unwanted longer
exon Antisense ? shorter isoform
d
Antisense-induced skipping
Nonsense mutation
x
Expendable exon (e.g., protein with many repeated
domains) Exon must be multiple of 3 in length to
maintain reading frame afterskipping
3
Sazani P, Gemignani F, Kang SH, Maier MA,
Manoharan M, Persmark M, Bortner D, Kole
R. Systemically delivered antisense oligomers
upregulate gene expression in mouse tissues. Nat
Biotechnol. 2002 Dec20(12)1228-33.
EGFP Enhanced green fluorescent protein model
system
Mutant globin intron has activated splice sites
Antisense RNA injected into tail vein, RNA was
modified for stability
Actin promoter, universally expressed.
Exon skipping yields green fluorescence
(HeLa cells)
4
RNA modification for stabilization
Morpholino instead of
Instead of deoxyribose or ribose
Modified phosphate
Still base pairs OK
5
Even more extreme and more stable RNA
modification peptide nucleic acids
B a nucleic acid base
Amide bonds, No ribose
PNA peptide nucleic acid
Attached 1 to 4 lysines here
Base pairs even better than natural nucleic acids
(higher melting temperatures)
6
RNA modification
Also can add 2 MOE
-O-CH2-CH2-O-CH3
methoxyethyl -
Phosphorothioate deoxyoligonucleotides
7
Splicing inhibition after injection of modified
RNAs into the mouse
8
Examples of therapeutic intervention at the level
of pre-mRNA splicing
- Interfere with improper splicing caused by splice
site creation or activation - E.g., beta-thalassemia (R. Kole) in which a
splice site has been created by a mutation - Use complementary DNA (antisense)
- Rapidly degraded Use modified bases, sugars
PNA, morpholino, 2 OMe, - Normally, DNA-RNA hybrids endogenous RNase H
type activity? RNA destruction - Modified antisense DNA circumvents this problem
(dont want mRNA destroyed here, want to correct
its splicing
B. Bias alternative splicing ratios
Target the unwanted isoform exon-intron
joint. e.g., BCL-2 isoforms, one is
pro-apoptotic, one anti-apoptotic. The latter
increased in many cancersTarget the
anti-apoptotic isoform in cancer cells. e.g.,
GABA-a-gamma-2 (GABA gamma amino butyric acid,
a neurotransmitter) receptor Long and short
forms. Long form associated with mental illness.
C. Skip offensive exons e.g., nonsense
truncations in dystrophin
9
Dystrophin gene 2400 kb, mRNA 14 kb, 79 exons
a giant gene Protein maintains muscle cell
membrane integrity Mutation Duchennes muscular
dystrophy Some (half) due to stop codon
(nonsense) in a repetitious exon (spectrin-like
repeat) Deliver antisense to ends of exon with
the nonsense mutation in mdx mice (model for
Duchennes). Use AAV (adeno-associated virus) to
deliver the antisense gene Measure mRNA with
skipped exon Dystrophin protein muscle
histochemistry for dystrophin
10
Use antisense RNA to target the branch point
upstream of the offending exon 23 and the donor
splice site downstream of the exon.
protein
mRNA
(nonsense mutation)
3 X 71, so skipping ? no
frameshift
79
SD splice donor
Branch site (consensus YNYTRAY)
BP branch point
Sequences targets by antisense
11
Adeno associated virus construct for splicing
inhibition
U7 promoter
ITR inverted terminal repeat(for replication
and packaging)
Consensus binding site for Sm proteins (to
target to pre-mRNA)
compl. to splice donor site
compl. to branch
Double target synergistic (loop?) (Kole)
U7 normally hybridizes with seq. at 3 end of
histone mRNAs to effect cleavage Binds 2 Sm
proteins in coiled (Cajal) bodies (RNA
processing centers?) low concentrations (1000s
of molecules per cell) U7OPT Change Sm binding
site to consensus for all snRNAs (spliceosomal,
for delivery there) high copy no. no longer in
coiled bodies Sm proteins common to many
small ribonucleoproteinss used in RNA
processing Now include anti-splice site segments
as well. In permanent transfectants can effect gt
50 inactivation of a globin cryptic
site. Gorman, L., Suter, D., Emerick, V.,
Schumperli, D. Kole, R.. Proc Natl Acad Sci U S
A 95, 4929-34 (1998).
12
Exon skipping induced about 2 weeks after
injection
Expression of U7 antisense construct
RT-PCR
U7OPT-A.S.
Endog. U7
(slow onset conclude slow mRNA turnover)
0 2 4 6 13 weeks
sm size markers
Splicing assay (RT-PCR)
Skip exon 23, after 2-4 wks.
0 2 4 6 8
13 weeks
normal
Dystrophin protein (Western)
13
dystrophin
dystrophin-associated antigens
Muscle immuno-histochemistry
Normal
Untreated mdx
Treated mdx
Top, middle ,and bottom
14
Nucleic acid aptamers Aptamers molecules that
bind other molecules with good affinity and
specificity Usually these are proteins But they
can also be RNA or DNA. That is, single stranded
RNA or DNA molecules can and will fold up into
secondary and tertiary structures depending on
their sequence. DNA can be synthesized as very
large numbers of different (random sequences)
Aptamers can be selected from among these
molecules based on their ability to bind an
immobilized ligand. The tiny fraction found by
chance to be able to bind to your favorite ligand
can by amplified by PCR (along with background
molecules). Re-iteration of the procedure will
enrich for the aptamer until they dominate the
population. At this point they can be cloned and
sequenced. RNA molecules can be selected by
synthesizing them from a randomized DNA
population using the T7 promoter appended to each
DNA molecule. This enrichment procedure is just
the SELEX method described earlier for finding
the RNA substrate for RNA binding proteins. In
this case its the same procedure, looked from
the opposite point of view not what RNA will the
protein bind best, but what RNA binds the protein
best.
15
SELEX Have a random 40-mer synthesized, between
2 arbitrary 20-mers (PCR sites) 440
1024 Practical limit 1015 2 nmoles
50 ug DNA 1015 is a large number.Very
large (e.g., 500,000 times as many as all the
unique 40-mers in the human genome. These 1015
sequences are known as sequence space Each DNA
molecule of these 1015 (or RNA molecule copied
from them) can fold into a particular 3-D
structure. We know little as yet about these
structures. But we can select the molecules that
bind to our target by AFFINITY CHROMATOGRAPHY
20-mer
Random 40
20-mer
16
Previously discussed SELEX in terms of finding
the substrate sequence(s) for an RNA binding
protein.Here select an RNA sequence that can
bind any target if interest (protein, small
molecule)
SELEX Systematic Evolution of Ligands by
EXponential enrichment for RNA
Essential elements1) Synthesis of randomized
DNA sequences 2) In vitro T7 mediated
RNAsytnhesis from DNA 3) Afinity
chromatography 4) RTPCR
(1015)
RNA
Ligand is immobilized here. Small molecule or
large molecule.
DNA
RNA
RNA
17
Some examples of aptamer targets Zn2 ATP adenosin
e cyclic AMP GDP FMN (and an RNA aptamer is
found naturally in E.coli) cocaine dopamine amino
acids (arginine) porphyrin biotin organic dyes
(cibacron blue, malachite green) neutral
disaccharides (cellobiose) oligopeptides aminoglyc
oside antibiotics (tobramycin) proteins
(thrombin, tat, rev, Factor IX, VEGF, PDGF,
ricin) large glycoproteins such as CD4 anthrax
spores (?)
18
Tobramycin (antibiotic) bound to its aptamer
(backbone)
Streptomycin-binding aptamer
Teriary structure annotations
19
Tobramycin (antibiotic) bound to its aptamer
(backbone)
20
theophilline
FMN
AMP
Hermann, T. and Patel, D.J.2000. Adaptive
recognition by nucleic acid aptamers. Science
287 820-825.
theophilline
FMN
Aromatic ringstacking interactions
RNA
RNA
AMP
AMP
H-bonding
RNA
DNA
Specificity Caffeine theophilline a methyl
group on a ring N (circle) bindingis gt1000
times weaker
Orange ligand Blue RNA functional
groups Green planar surface for stacking
interactions
21
Electrostatic surface mapred - blue
Base flap shuts door
22
Hermann, T. and Patel, D.J.2000. Adaptive
recognition by nucleic acid aptamers. Science
287 820-825.
MS2 protein as beta sheet bound via protruding
side chains
23
G-quartets dominate the structure of
anti-thrombin DNA aptamers
Note Target is a protein Aptamer can be DNA not
RNA)
An example of a tertiary structure motif
24
Stabilizing RNA aptamers RNA aptamers are
unstable in vivo (bloodstream) DNA aptamers are
more stable but still can be destroyed by DNases.
Modification to protect 2 F-YTP
(Y pyrimidine) 2 NH2-YTP But not
substrates for PCR enzymes. But yes OK for T7 RNA
polymerase and reverse transcriptase. So
Isolation of an RNase-resistant aptamer
1015
random
DNA synthesizer
PCR site
T7 prom
T7 polymerase, 2F-CTP 2F-UTP
2F-RNA
Lots of normal DNA version
Affinity chromatography selection
PCR
Enriched stableaptamer
Reverse transcriptase
Normal DNA version
Normal deoxynucleoside triphosphates
Final product after N iterations
25
Aptamer vs. prostate cancer cell membrane antigen
(PMSA), conjugated to rhodamine
Lupold, S.E., Hicke, B.J., Lin, Y., and Coffey,
D.S. 2002. Identification and characterization
of nuclease-stabilized RNA molecules that bind
human prostate cancer cells via the
prostate-specific membrane antigen. Cancer Res
62 4029-4033.
Potential use as an anticancer diagnostic, and
therapeutic.
26
Therapeutic use of an aptamer that binds to and
inhibits clotting factor IX
Rusconi, C.P., Scardino, E., Layzer, J., Pitoc,
G.A., Ortel, T.L., Monroe, D., and Sullenger,
B.A. 2002. RNA aptamers as reversible
antagonists of coagulation factor IXa. Nature
419 90-94.
Reading
Facytor IX acts together with Factor VIIIa to
cleave Factor X, thus activating it in a step in
the blood coagulation cascade leading to a
clot. Thus inhibiton of Factor IX results in
inhibition of clot formation. Dessirable during
an angioplasty, for example. The usual
anti-coagulant used in angiplasty is heparin,
which has some toxicitiy and is difficult to
control. .
Inverted T at 3 end (3-3) slows exonucleolytic
degradation ( R-3O-P-O-3-R-T )
27
Anti-Factor IX RNA aptamer isolated by SELEX
Kd for Factor IX 0.6 nM
FIXa FVIIIa cleave FX
Aptamer inhibits this activity
aptamer _/- PEGylation
Clotting time increase
Mutant version
-aptamer 1
Conjugate to polyethylenglycol to increase
bloodstream lifetime
PEG polyethyleneglycol polymer, appended to
decrease clearance rate.
28
An antidote to stop the anti-clotting action if a
patient begins to bleed Just use the
complementary strand (partial). The 2 strands
find each other in the bloodstream!
Antidote 5-2 design open squares
16-fold excess
duplexed
In human plasma
free aptamer
Anti-coagulant activity
Scrambled antidote
Oligomer 5-2
Ratio of anti- to aptamer
29
Need 10X antidote
Antithrombin aptamer antidote tested in human
serum
Antidote lasts a long time
30
Reduced clotting
Reversed by antidote
In serum of patients with heparin-induced
thrombocytopenia (can no longer use heparin)
31
Macugen an RNA aptamer that binds VEGF and is
marketed for adult macular degeneration (wet type)
From the label
R
Where R is and n is
approximately 450. The chemical name for
pegaptanib sodium is as follows RNA,
((2'-deoxy-2'-fluoro)C-Gm-Gm-A-A-(2'-deoxy-2'-flu
oro)U-(2'-deoxy-2'-fluoro)C-Am-Gm-(2'-deoxy-2'-flu
oro)U-Gm-Am-Am-(2'-deoxy-2'-fluoro)U-Gm-(2'-deoxy-
2'-fluoro)C-(2'-deoxy-2'-fluoro)U-(2'-deoxy-2'-flu
oro)U-Am-(2'-deoxy-2'-fluoro)U-Am-(2'-deoxy-2'-flu
oro)C-Am-(2'-deoxy-2'-fluoro)U-(2'-deoxy-2'-fluoro
)C-(2'-deoxy-2'-fluoro)C-Gm-(3'?3')-dT), 5'-ester
with a,a'-4,12-dioxo-6-5-(phosphoonoxy)pentyl
aminocarbonyl-3,13-dioxa-5,11-diaza-1,15-pentade
canediylbis?- methoxypoly(oxy-1,2-ethanediyl),
sodium salt. The molecular formula for
pegaptanib sodium is C294H342F13N107Na28O188P28C2
H4On (where n is approximately 900) and the
molecular weight is approximately 50
kilodaltons. Macugen is formulated to have an
osmolality of 280-360 mOsm/Kg, and a pH of 67.
32
DNA aptamers for protein diagnostics
Photoaptamers, from Somalogic, Inc.
ORIGINAL SELEX PAPERC. Tuerk and L. Gold.
"Systematic evolution of ligands by exponential
enrichment RNA ligands to bacteriophage T4 DNA
polymerase," Science, 249505-10, 1990
Make DNA aptamers using bromouracil instead of
thymine Bromouracil absorbs near UV light (313
nm) (Normal DNA absorbs at 260 nm) Apply blood
to a photo-aptamer macroarray
Wash stringently toproduce a low
background (e.g., NaOH). Stain with a
fluorescent stainagainst proteins (e.g, for
primary amine groups)
albumin
prolactin
LDH
protein
B bromouracil bases
B
B
covalentcross-links
B
Different aptamers (many molecules per spot)
Low background afforded by stringent wash made
possible by covalent X-linking yields an
important increase in sensitivity
33
Ribozymes 1982 Cech
Tetrahymena rRNA intron is self-spliced out
(Guanosine GR Mg) Altman and Pace
Ribonuclease P RNP RNA component alone can
process the 5 ends of tRNAs Mitochondrial group
I introns (GR catalyzed) also can
self-splice Then group II introns in
mitochondria (lariat-formers) Mutations (100s)
revealed Internal guide sequence GR-binding
site secondary structure Conserved base
analysis (100s) ? confirms structure X-ray
diffraction a few 3-D structures
34
(natural ribozymes)
Free guanosine
No lariat
lariat
35
Another natural ribozyme Hammerhead ribozymes
(self-cleavage) plant viroids and human delta
virus (with Hepatitis C)
Self-cleavage is via the hammerhead motif
36
Hammerhead ribozyme(RNase) can cleave in
trans (hammer head is upside down)
Synthetic variation(cleaves in trans)
You are in charge of what it will cleave
37
Model of hammerhead ribozyme (data based)
38
New synthetic ribozymes, and DNAzymes Start with
1015 DNA molecules again Select for enzyme
activity E.g., cleaves itself off a solid
support in the presence of Mg Many different
activities have been selected.Most have to do
with nucleic acid transformationsRNase, ligase,
kinase, etc.But not all (C-C bond formation
possible). Generally much slower than protein
enzymes. Most work has been on RNases (usually
associated with the word ribozymes)
39
You can use SELEX to isolate new artificial
ribozymes
Tang, J. and Breaker, R.R. 2000. Structural
diversity of self-cleaving ribozymes. Proc Natl
Acad Sci U S A 97 5784-5789.
1015 DNA molecules with T7 promoter
Keep molecules under non-permissive conditions
so they stay intact (without Mg)
Proposedcleavage zone
1. RT -gt cDNA Zone is rebuilt by being part of
the primer PCR lots of DS-DNA 2. T7
transcription-gt Lots of RNA
Now add Mg
Selecting for cleavage anywhere in the zone
Proposedcleavage zone
i.e., al 16 dinucleotides present as possible
cleavage sites
40
12 different in vitro evolved ribozyme structures
Tang, J. and Breaker, R.R. 2000. Structural
diversity of self-cleaving ribozymes. Proc Natl
Acad Sci U S A 97 5784-5789.
Hammerhead was one
Common substrate (all 16 dinucleotide combos)
Rate (min-1)
41
Combine an aptamer and a ribozyme ? Allosteric
ribozyme Catalytic activity can be controlled by
ligand binding! Positive or negative. Modular M
olecular switches, biosensors
42
Isolation of aptamer-ribozyme combinations That
respond to ligand binding.
Selection of an allosterically activated ribozyme
Iterations
Select with decreasing activation times for beter
and better binders.
Selection of an allosterically inhibited ribozyme
Soukup, G.A. and Breaker, R.R. 1999. Engineering
precision RNA molecular switches. Proc Natl Acad
Sci U S A 96 3584-3589.
43
The same induction communication module can be
used with several different allosteric aptamer
modules
FMN responsive
Theo responsive
ATP responsive
k rate of catalysis
With ligand Without ligand
Soukup, G.A. and Breaker, R.R. 1999. Engineering
precision RNA molecular switches. Proc Natl Acad
Sci U S A 96 3584-3589.
44
Using an allosteric ribozyme to create a chemical
sensor
Reading
Frauendorf, C. and Jaschke, A. 2001. Detection
of small organic analytes by fluorescing
molecular switches. Bioorg Med Chem 9 2521-2524.
Start with a theophylline-dependent ribozyme
Analogy A molecular beacon that respond to
nucleic acid hybridization
45
Frauendorf, C. and Jaschke, A. 2001. Detection
of small organic analytes by fluorescing
molecular switches. Bioorg Med Chem 9 2521-2524.
Too short to maintain a stable duplex structure
Separate substrate molecule (in trans),
fluorescently tagged
Nearby quenching group
46
H
theophylline
5X effect
caffeine
good specificity
Not so sensitive (0.3 mM)
47
Prominent aptamer companies Archemix
(Boston) Somalogic (Colorado) Noxxon (Germany)
48
Extra topic graphics
49
Winkler, W., Nahvi, A., and Breaker, R.R. 2002.
Thiamine derivatives bind messenger RNAs
directly to regulate bacterial gene expression.
Nature 419 952-956.
Back to Nature Aptamers play a role in
regulation of gene expression
Thiamine Inhibits its own synthesis (in
bacteria)
50
- thi Translation takes place
Shine-Delgarno sequenceribosome binding site to
initiate translation
5 end ofthiM mRNA
51
DNA can also form enzymes DNAzymes
Li, Y. and R. R. Breaker (1999).
"Deoxyribozymes new players in the ancient
game of biocatalysis." Curr Opin Struct Biol
9(3) 315-23.
Selection scheme for self-cleaving DNase DNAzymes
Putative cleavage region
biotin
Solid phase streptavidin
Selecting for DNAzymes that will only cleave in
the presence of the cofactor (otherwise would
self-destruct)
Pb and Cu have been described
Collect freed large fragment
PCR with large biotinylatedleft primer that
reconstructs cleavage site(not part of the
random region)
52
Emilsson, G. M. and R. R. Breaker (2002).
Deoxyribozymes new activities and new
applications.Cell Mol Life Sci 59(4) 596-607.
Some DNAzyme activities
Compare protein enzymes, Typically 6000 on this
scale (100/sec)
53
Selection scheme for an allosteric (regulatable)
self-cleaving DNase DNAzyme
SELECTION STEP
Ligand binding domain
Communication domain
Catakytic domain
DNAzyme will only cleavein the presence of the
cofactor (for activator) or in the absence of
the Cofactor (for inhibitor)
54
An increasing number of DNAzyme activities are
being isolated Ligase Polymerase DNase And
activities using co-enzymes, as protein enzymes
do E.g., co-enzyme A