Automated Refinement PowerPoint PPT Presentation
Title: Automated Refinement
1
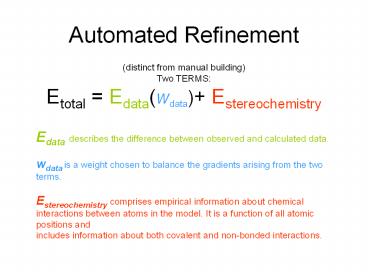
Automated Refinement
(distinct from manual building) Two
TERMS Etotal Edata(wdata) Estereochemistry Ed
ata describes the difference between observed and
calculated data. wdata is a weight chosen to
balance the gradients arising from the two
terms. Estereochemistry comprises empirical
information about chemical interactions between
atoms in the model. It is a function of all
atomic positions and includes information about
both covalent and non-bonded interactions.
2
Edata (R-factor)
positive negative density density
Move atoms to minimize the R-factor. Discrepancy
between Fobs and Fcalc. Specifically, minimize
E ES w(Fobs-Fcalc)2 Over all hkl. Least squares
refinement. Atoms shift toward positive density
in a difference Fourier electron density
map. r(x)1/VSFobs-Fcalce-2pi(hx-fcalc)
Radius of convergence is limited
3
Estereochemistry (Geometry)
- BOND LENGTHS ANGLES have standard values. Engh
Huber dictionary. - - CHIRALITY of a-carbons
- PLANARITY of peptide bonds and aromatic side
chains - NONBONDED CONTACTS -two atoms cannot occupy the
same space at the same time - TORSION ANGLE PREFERENCES side chains have
preferred rotamers. - some values of f and y are forbidden.
-Ramachandran. Not restrained- used for
validation.
4
Jeopardy clueThe appearance of the atomic
model when stereochemical restraints are not
included in crystallographic refinement.
Etotal Estereochemistry wdataEdata
What is spaghetti, Alex?
5
Importance of supplementing the Data to
Parameter Ratio in crystallographic refinement.
PARAMETERS Each atom has 4 parameters (variables)
to refine x coordinate y coordinate z
coordinate B factor In proteinase K there are
approximately 2000 atoms to refine. This
corresponds to 20004 8000 variables.
DATA At 2.5 A resolution we have 8400
observations (data points) (Fobs). When of
observations of variables A perfect fit can be
obtained irrespective of the accuracy of the
model.
At 1.7 A resolution we have 25,000
observations. About 3 observations per variable.
The reliability of the model is still
questionable.
Adding stereochemical restraints is equivalent to
adding observations
6
2nd Jeopardy clueThe value of the R-factor
resulting when stereochemical restraints are not
included in crystallographic refinement.
Etotal Estereochemistry wdataEdata
What is zero, Alex?
7
An atomic model should be validated by several
unbiased indicators
therefore
Rfree is an unbiased indicator of the discrepancy
between the model and the data. The data used in
this R-factor calculation were not used in
determining atomic shifts in the refinement
process. Ramachandran plot is unbiased because
phi and psi torsion angles are not restrained in
the refinement process.
8
BAD
2.8 Å
H
O
N
H
Asn
9
GOOD
2.8 Å
H
O
N
H
Asn
10
ERRAT plot examines the geometric relationship
between non-bonded atoms. Looks at the fraction
of non-bonded contacts with C, N, O as a function
of distance.
11
Verify 3D plot Gives an indication if the
sequence has been improperly threaded through the
backbones. Each of the 20 amino acid types has a
characteristic (1) Surface area buried (2)
fraction of side-chain area covered by polar
atoms (3) local secondary structure. Verify 3D
plots correlation between ideal and your model.
Compatibility of a model with its sequence.
RuBisCo chain traced backwards
12
Plan for today
- The refinement of native proteinase K is assumed
to be complete thanks to ARP/wARP. - You will refine the structure of the proteinase
K- PMSF complex using the Fobs data measured
earlier in the course. - The starting model for the refinement will be the
native proteinase K structure. - Begin 5 cycles of automated refinement. This
will only move atoms. It will not add new atoms. - Then manually build the PMSF inhibitor into an
Fo-Fc difference Fourier map. Refinement process
typically iterates between automated and manual
building. Automated refinement has a limited
radius of convergence. For example- automated
refinement cannot jump between rotamers or flip
between cis and trans peptides. - Validate structure. Fill out Refinement
Statistics table.
13
Difference Fourier map
r(x)1/VSFobs-Fcalce-2pi(hx-fcalc)
Here, Fobs will correspond to the
ProteinaseK-PMSF complex. Fcalc will correspond
to the model of Proteinase K by itself after a
few cycles of automated refinement. Positive
electron density will correspond to features
present in the PMSF Complex that are not in the
native structure. Negative electron density will
correspond to features present in the native
structure that should be removed in the inhibitor
complex. After model building, do more automated
refinement and then validate.
14
Cis vs. Trans peptide
15
Cis OK with glycine or proline
O
peptide plane
C
N
Ca
Ca
R
Steric hindrance equivalent for cis or trans.
16
Steric hindrance equivalentfor cis or trans
proline
O
peptide plane
Ca
Cd
Cb
Cg
C
N
Cg
Cb
Cd
Ca
R
.
17
Name _______________________
Proteinase K PMSF complex