Multiple Sequence Alignment - PowerPoint PPT Presentation
1 / 39
Title: Multiple Sequence Alignment
1
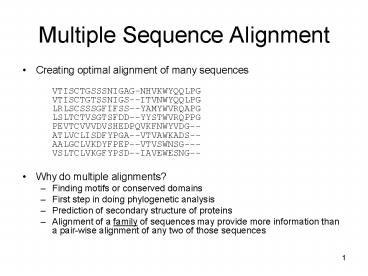
Multiple Sequence Alignment
- Creating optimal alignment of many sequences
- VTISCTGSSSNIGAG-NHVKWYQQLPG VTISCTGTSSNIGS--ITVN
WYQQLPG LRLSCSSSGFIFSS--YAMYWVRQAPG
LSLTCTVSGTSFDD--YYSTWVRQPPG PEVTCVVVDVSHEDPQVKFN
WYVDG-- ATLVCLISDFYPGA--VTVAWKADS--AALGCLVKDYFPE
P--VTVSWNSG--- VSLTCLVKGFYPSD--IAVEWESNG-- - Why do multiple alignments?
- Finding motifs or conserved domains
- First step in doing phylogenetic analysis
- Prediction of secondary structure of proteins
- Alignment of a family of sequences may provide
more information than a pair-wise alignment of
any two of those sequences
2
Pairwise vs Multiple
- Pairwise
- Used to identify previously unknown biological
relationship based on sequence similarity - Multiple
- Inverse of pairwise
- Based on known biological relationships between
sequences, identify unknown conserved subpatterns
3
Multiple Sequence Alignment
- For pair-wise alignment
- Dynamic Programming
- Heuristic
- Whats the difference?
- Which one makes sense to model after?
4
Approximation Methods
- Progressive
- Iterative
- Locally Conserved Pattern
- Statistical and Probabilistic
5
Progressive Global Alignment
- Uses dynamic programming
- of sequences is small for reason of DP
- For pair-wise
- Assume 2 protein sequences of length 300
- of comparisons?
- For multiple
- Assume 3 sequences of length 300
- of comparisons?
- Assume 4 sequences,
- of comparisons?
3002 9x104
3003 2.7x107
3004 8.1x109
6
(No Transcript)
7
(No Transcript)
8
Scoring a Multiple Alignment
- Let
- A be a finite alphabet
- for DNA, A A, C, G, T
- for AA, A set of all 20 amino acids
- A A U -
- a1,,ak be k sequences over A
- Assume each string contains n characters
9
Scoring a Multiple Alignment
- Alignment of ak sequences is a dimensional
matrix, M - Each element of M is a member of A
- Each row i contains characters of ai (and a
possible gap -) - Every column contains at least one symbol from A
10
Scoring a Multiple Alignment - Sum of Pairs
- A12 Alignment score of sequence 1 and sequence
2 - A13 Alignment score of sequence 1 and sequence
3 - A23 Alignment score of sequence 2 and sequence
3 - OAij Optimal alignment score of sequence i and
j - Alignment Divergence eij Aij OA(ij)
- Degree of Divergence d ?e
- The larger eij the more divergent the msa from
the pair-wise alignment gt smaller contribution
to the MSA - Closely related sequences will have low
divergence - Distantly related sequences will have high
divergence
11
Progressive Alignment
- ClustalW
- Uses a heuristic alignment approach
- Build a multiple alignment progressively by a
series of pair-wise alignments - Align most closely related sequences gradually
adding in more distant ones - Known as a greedy algorithm
12
Problems with Progressive Alignment
- Local Minimum
- Dependence of final MSA on initial pair-wise
alignments (incorrect branching order in initial
tree) - Highly divergent sequences (lt30 identity) causes
progressive approach to be much less reliable - Parameter Choice
- Choice of suitable scoring matrices and gap
penalties (different matrices are optimal at
different evolutionary distances) - Range of gap penalties, will find correct or best
possible solution, can be very broad of highly
similar sequences
13
ClustalW
- Basic Algorithm
- Align all pairs of sequences to calculate
distance matrix - Calculate guide tree from distance matrix
- Progressively align sequences according to
branching order in guide tree
14
Distance Matrix / Pairwise Alignments
- Fast Approximate Method
- Heuristic
- Scores calculated as
- of k-tuples matches between two sequences
(gap penalty of gaps) - k1,2 for aa, 2-4 for dna
- Slow Accurate Method
- Dynamic Programming
- Score
- 1 (( of identities / length of sequences) /
100))
15
Guide Tree
- ClustalW initially used UPGMA
- Unweighted Pair Group Method by Arithmetic Mean
- Simplest method of tree construction
- Assumes equal rates of mutation along the
branches - UPGMA Algorithm
- Definition Node in a tree is called an
Operational Taxonomic Unit (OTU) - From distance matrix, cluster pair of OTUs with
smallest distance, and calculate new distance - Repeat previous step until clusters converge
16
Guide Tree - UPGMA
- Cluster pair with smallest distance
- Recalculate distance matrix
17
Guide Tree - UPGMA
- Calculate new distance using composite OTU(A,B)
- Distance between a simple OTU and a composite OTU
is the average of the distances between the
simple OTU and the constituent simple OTUs of the
composite OTU - dist (A,B),C (dist A,C dist B,C) / 2 (4
4) / 2 4dist (A,B),D (dist A,D dist B,D) /
2 (6 6) / 2 6dist (A,B),E (dist A,E
dist B,E) / 2 (6 6) / 2 6 dist (A,B),F
(dist A,F dist B,F) / 2 (8 8) / 2 8
18
Guide Tree - UPGMA
- Calculate new distance using composite OTU(A,B)
- Distance between a simple OTU and a composite OTU
is the average of the distances between the
simple OTU and the constituent simple OTUs of the
composite OTU
19
Guide Tree - UPGMA
- Second Iteration
20
Guide Tree - UPGMA
- Third Iteration
21
Guide Tree - UPGMA
- Fourth Iteration
22
Guide Tree - UPGMA
- Fifth Iteration
23
Guide Tree
- ClustalW uses Neighbor-Joining
- Assumes unequal rates of mutation along each
branch - Produces tree with branch lengths proportional to
estimated divergence along each branch - Neighbor-Joining Algorithm
- Find pairs of OTUs that minimize total branch
length at each stage of clustering starting with
a starlike tree (Minimum-Evolution Tree).
24
Guide Tree - Neighbor-Joining
- Start with a star tree with N nodes
- Combine the pair with the smallest branch lengths
- Continue until all N-3 interior branches are
found - Dij distance between OTUs i and j
8
1
7
X
6
2
3
5
4
25
Definitions
- Lab branch lengths between nodes a and b
- Sum of branch lengths
8
1
7
X
6
2
3
5
4
26
Definitions
- Assuming 1 2 are any pair of (closest)
neighbors - Any pair of OTUs can take the position of 1
2, N(N-1)/2 waysof choosing pairs - Choose the pair that gives the smallest branch
lengths
1
8
7
X
2
Y
6
3
5
4
27
Definitions
- Branch lengtbetween XY is now
- Removing XY givestwo star-like trees
1
8
7
X
2
Y
6
3
5
4
28
Definitions
- Sum of branch lengths
- If 12 are closestneighbors, join themto make
new OTU and recalculate distance
1
8
7
X
2
Y
6
3
5
4
29
Definitions
- To find the tree branch lengths when 3 nodes left
1
8
7
X
2
Y
6
3
5
4
30
Guide Tree - Neighbor-Joining
- Calculate each branch length
8
1
7
X
6
2
3
5
4
31
Guide Tree - Neighbor-Joining
- Calculate each branch length
8
1
7
X
6
2
3
5
4
32
Guide Tree - Neighbor-Joining
- Calculate each branch length
1
8
7
X
2
Y
6
3
5
4
33
Guide Tree - Neighbor-Joining
- Calculate each branch length
1
8
7
2
X
6
3
5
4
34
Guide Tree - Neighbor-Joining
- Recalculate distances
- Recalculate sum of branch lengths
1
8
7
2
X
6
3
5
4
35
Guide Tree - Neighbor-Joining
- Start next iteration, nodes 5 6
1
8
7
2
X
6
3
5
4
36
Guide Tree - Neighbor-Joining
1
8
7
2
X
6
3
5
4
37
Guide Tree - Neighbor-Joining
- Next Iteration (1-2) 3
1
8
7
2
X
6
3
5
4
38
Guide Tree - Neighbor-Joining
8
1
7
2
Y
X
6
3
5
4
39
Guide Tree - Neighbor-Joining
8
1
7
2
X
6
3
5
4
40
Progressive Alignment
- Use a series of pairwise alignments to align
larger and larger groups of sequences, following
the branching order in the guide tree - Align the most closely related sequence then add
the next more closely related sequence,
iteratively - Full DP algorithm is used by aligning two
existing alignments or sequences - Gaps in present/older alignments remain fixed