Chapter 3: Amino Acids - PowerPoint PPT Presentation
1 / 144
Title:
Chapter 3: Amino Acids
Description:
the structures of the side chains of all 20 natural amino acids ... The protein is put in a bag of cellulose membranes having small pores of controlled size. ... – PowerPoint PPT presentation
Number of Views:152
Avg rating:3.0/5.0
Title: Chapter 3: Amino Acids
1
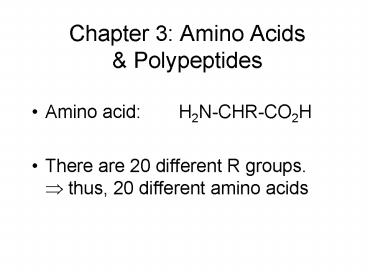
Chapter 3 Amino Acids Polypeptides
- Amino acid H2N-CHR-CO2H
- There are 20 different R groups.? thus, 20
different amino acids
2
Zwitterion
- In aqueous solution, the amino and carboxylic
acid groups will ionize to give the zwitterionic
formH3N-CHR-CO2-
3
(No Transcript)
4
Stereochemistry
- Note that the R group means that the ?-carbon is
a chiral center. - All natural amino acids are L-amino acids.
- This means that almost all have the S
configuration. (Exceptions glycine and cysteine
can you tell why?)
5
(No Transcript)
6
(No Transcript)
7
(No Transcript)
8
(No Transcript)
9
(No Transcript)
10
(No Transcript)
11
(No Transcript)
12
(No Transcript)
13
(No Transcript)
14
(No Transcript)
15
You should know
- the structures of the side chains of all 20
natural amino acids - both the 1-letter and 3-letter codes (see Table
3-1). - the pKa's of the 7 ionizable R groups (only to 1
decimal place) - Although you don't have to memorize all of the
pKa's for every carboxylate and amine, you should
know that they are all 2 and 9-10, respectively
(with a few exceptions). You should should be
able to calculate pI if given the pKa's.
16
(No Transcript)
17
(No Transcript)
18
(No Transcript)
19
Can group into several categories
- 1) alkanes A V I L (P)
- 2) aromatics F W Y (H)
- 3) carboxylates D E
- 4) corresponding amides N Q
- 5) positively charged K R (H)
- 6) Sulfur-containing C M
- 7) hydroxyls S T Y
20
Can group into several categories
- 8) ?-branched V T I
- 9) small G A S C (V T)
- 10) large W R Y F (M)
- 11) H-bond donors S T Y N Q K R H W (D E, if
protonated) - 12) H-bond acceptors S T Y N Q D E R H W
21
How to calculate pI
- The isoelectric point (pI) of an amino acid or
peptide is the pH at which the charge of the
molecule 0. - It can be calculated simply as the arithmetic
mean of the 2 pKa's corresponding to the
transitions generating the 1 and -1 forms.
22
How to calculate pI
- Heres how to do it
- Identify all ionizable groups
- Assign pKas to each ionizable group
- Start with each ionizable group in protonated
form (very low pH maybe 0 or 1) and calculate
its net charge - Slowly move up in pH to the first ionizable
groups pKa and deprotonate it (reduce charge by
1) - Do this until each group is deprotonated. Now
you have identified all charged forms and at
which pH each transition occurs. - Identify the form with net charge 0
- Take the pKa on either side of the electrically
neutral form and take their average. This is the
pI.
23
How to calculate pI
- Take Glycine as an example it has only 2
ionizable groups. The transition (from low to
high pH) would beGly1 ? Gly0 ? Gly -1 - pKa (-CO2H) 2.34 pKa (-NH3) 9.60
- pI (2.34 9.60)/2 11.94/2 5.97
24
(No Transcript)
25
How to calculate pI
- Glutamate has an ionizable group (-CO2H pKa
4.25) that generates a negative charge when
deprotonated.Its transitions would be - Glu1 ? Glu0 ? Glu-1 ? Glu-2
- The relevant pKa 's are
- pKa(-CO2H) 2.19 pKa(R) 4.25
- pI (2.19 4.25)/2 6.44/2 3.22
26
(No Transcript)
27
How to calculate pI
- Histidine has an ionizable group (imidazole pKa
6.00) that is positively charged when
protonated. Its transitions would be - His2 ? His1 ? His0 ? His-1
- The relevant pKa 's are
- pKa(R) 6.00 pKa(-NH3) 9.17
- pI (6.00 9.17)/2 15.17/2 7.59
28
(No Transcript)
29
Making dipeptides
- H3N-CHR-CO2- H3N-CHR-CO2- ?
- H3N-CHR-CONH-CHR-CO2- H2O
- This process can be repeated to make a tripeptide
and so on - H3N-CHR-CONH-CHR-CO2- H3N-CHR-CO2- ?
H3N-CHR-CONH-CHR-CONH-CHR-CO2-
30
(No Transcript)
31
(No Transcript)
32
Making dipeptides
- The C-N bond has partial double bond character,
making the -CONH- moiety planar. This limits the
orientations available to the polypeptide - (e.g. the barrier of rotation about the C-N bond
in formamide is 18 kcal/mol)
33
..
34
(No Transcript)
35
(No Transcript)
36
Hydrolysis of polypeptides amino acid analysis
- Polypeptides can be hydrolyzed to constituent
amino acids. - This is typically done by boiling the polypeptide
in 6 M HCl for 24 hours. - H3N-CHR-CONH-CHR-CONH-CHR-CO2- 2 H2O ? 3
H3N-CHR-CO2-
37
Hydrolysis of polypeptides amino acid analysis
- The R groups remain intact, except for
- Trp indole ring damaged
- Asn, Gln converted to Asp, Glu
38
Hydrolysis of polypeptides amino acid analysis
- The amino acids can be derivatized with
o-phthalaldehyde to make fluorescent derivatives
that are easy to detect. - These are chromatographed by reverse-phase HPLC
(high-pressure liquid chromatography). - The characteristic retention times are used to
identify the amino acids. - The fluorescence level can be quantified to
determine the amount of that amino acid.
39
Amino acid analysis.
40
Disulfide bonds
- 2 cysteine ? cystine
- 2 R-SH ? R-S-S-R (Note This is an oxidation)
- Intracellular conditions are maintained
sufficiently reducing to inhibit formation of
most disulfide bonds. - Extracellular conditions (as well as those found
in some organelles) are more oxidizing, favoring
disulfide formation. - Thus, extracellular proteins containing cysteines
often have disulfides, while intracellular
(cytosolic) proteins rarely have disulfides.
41
(No Transcript)
42
Reactions with amino acids
- I. Amino group
- Acylation ? R-(CO)-NH-R
- Ninhydrin reactionCauses oxidative
decarboxylation of ?-amino acids, and release of
ammonia, which reacts with a second molecule of
ninhydrin to form a purple product.(You dont
need to know details just know that it reacts
with any free amino group and the final product
is purple.)
43
Reactions with amino acids
- I. Amino group
- Ninhydrin reaction
44
Reactions with amino acids
- Fluorodinitrobenzene reaction
- Dansyl chloride reaction
45
Reactions with amino acids
- Fluorescamine reactionSimilar to dansyl chloride
forms fluorescent adduct.(Dont need to know
details.)
46
Reactions with amino acids
- o-phthalaldehyde reaction
- Schiff's base formation
- R-HCO NH2-R' ? R-HCN-R' H2O
- 8. Edman degradation (more later)
47
Reactions with amino acids
- Carboxyl group
- Amide formation
- Ester formation
- Acyl halide formation
- Reduction to alcohol (via aldehyde)
48
Reactions with amino acids
- Side chains
- R-SH (cysteine) R-S-S-R (cystine)
- Reduction of disulfide with ?-mercaptoethanolR-S-
S-R' HS-CH2CH2OH ?? R-S-S-CH2CH2OH
R'-SHR-S-S-CH2CH2OH HS-CH2CH2OH ?
HOCH2CH2-S-S-CH2CH2OH R-SH(driven by mass
action) - Can also accomplish with dithiothreitol (DTT)
(only requires 1 molecule to reduce 1 disulfide)
49
Reactions with amino acids
- Coupling to N-ethylmaleimide(blocks disulfide
formation after reduction) - Carboxymethylation with iodoacetate (introduces
carboxylate) R-SH I-CH2-COOH ?
R-S-CH2-COOH HI
50
Reactions with amino acids
- Reaction with ethyleneimine (introduces amino
group) - 5. Performic acid oxidation to cysteic acid
- 3 HCOOOH R-SH ? R-SO3H 3 HCOOH 5 HCOOOH
R-S-S-R H2O ? 2 R-SO3H 5 CHOOH - 6. Reaction with mercurials R-SH R'-Hg-Cl ?
R-S-Hg-R' HCl
51
Reactions with amino acids
- B. Imidazole (histidine)
- 1. Acetylation (with IAA)
- C. Phenol (tyrosine)
- Nitration
52
Reactions with amino acids
- Acetylation
- iodination (important use isotopic labeling
with 125I)
53
Reactions with amino acids
- D. ?-NH2 (lysine)
- Acylation R-NH-R
- Fluorodinitrobenzene reaction
- Dansyl chloride reaction
- o-phthalaldehyde reaction
- Schiff's base formation
- cyanylationR-NH2 HNCO ? R-NH-CONH2
- amidination
54
Reactions with amino acids
- R-COOH (glutamate, aspartate)
- Amidation
- Esterification
- Acyl halide formation
- Reduction to alcohol
- R-CONH2 (glutamine, asparagine)
- DeamidationR-CONH2 H2O ? R-COOH NH3
55
Protein Sequencing Strategy
- 1) Purify protein (methods discussed later)
- Cleave disulfides react with
- reducing agent followed by alkylating agent
- DTT or ?-ME
- NEM or IAA
- performic acid
56
(No Transcript)
57
Protein Sequencing Strategy
- Determine ends
- N-terminus react with FDNB or dansyl chloride
followed by acid hydrolysis and HPLC(Can also
perform Edman degradation with intact protein to
get N-terminal sequence, if the N-terminus is not
blocked.) - C-terminus digest with carboxypeptidase(not
often done)
58
Protein Sequencing Strategy
- Cleave polypeptide into smaller peptides
- A) Endopeptidases
- trypsin cleaves after Lys and Arg
- chymotrypsin cleaves after Phe, Tyr, or Trp
- endoproteinase Glu-C cleaves after Glu
59
(No Transcript)
60
Protein Sequencing Strategy
- Cleave polypeptide into smaller peptides
- B) Chemicals
- CNBr cleaves after Met(Lactone will be
hydrolyzed during acid hydrolysis.)
61
Protein Sequencing Strategy
- Purify peptides
- Characterize peptides(i.e. spectroscopy, amino
acid analysis, chromatography, etc.) - Most important determine amino acid sequence
by Edman degradation
62
(No Transcript)
63
Protein Sequencing Strategy
- 7) Reassemble sequence through overlaps of
peptides created by different means - 8) Map disulfides by cleaving protein into
peptides before disulfide bond cleavage. After
purification of disulfide-linked peptides and
cleavage of their disulfide bonds, sequencing of
the peptides should reveal which cysteines are
linked in disulfide bonds.
64
Edman degradation
- Q Why can you not use it to sequence long
polypeptides? - A Each step is completed with lt100 yield.
Eventually, less than half of the peptides have
the "current" amino terminal amino acid residue.
65
Example (inefficiency exagerated)
- H2N-V-D-R-G-T-H-K-L-S-F-E-W-Q-C-V-N-H2N-V-D-R-G-
T-H-K-L-S-F-E-W-Q-C-V-N-H2N-V-D-R-G-T-H-K-L-S-F-
E-W-Q-C-V-N-H2N-V-D-R-G-T-H-K-L-S-F-E-W-Q-C-V-N-
H2N-V-D-R-G-T-H-K-L-S-F-E-W-Q-C-V-N-H2N-V-D-R-
G-T-H-K-L-S-F-E-W-Q-C-V-N- - After 1st step
- H2N-D-R-G-T-H-K-L-S-F-E-W-Q-C-V-N-H2N-D-R-G-T-H-
K-L-S-F-E-W-Q-C-V-N-H2N-D-R-G-T-H-K-L-S-F-E-W-Q-
C-V-N-H2N-D-R-G-T-H-K-L-S-F-E-W-Q-C-V-N-H2N-D-
R-G-T-H-K-L-S-F-E-W-Q-C-V-N-H2N-V-D-R-G-T-H-K-L-
S-F-E-W-Q-C-V-N- - After 2nd step
- H2N-R-G-T-H-K-L-S-F-E-W-Q-C-V-N-H2N-R-G-T-H-K-L-
S-F-E-W-Q-C-V-N-H2N-R-G-T-H-K-L-S-F-E-W-Q-C-V-N-
H2N-R-G-T-H-K-L-S-F-E-W-Q-C-V-N-H2N-D-R-G-T-H-
K-L-S-F-E-W-Q-C-V-N-H2N-D-R-G-T-H-K-L-S-F-E-W-Q-
C-V-N- And so on
66
(No Transcript)
67
Synthesis of polypeptide
- Artificial (Merrifield process)
- Utilizes amino acid coupled to a solid phase
support through the carboxylate. - Amino acids with protected amino groups and
activated carboxylates react with the amino acid
to make N-terminally protected dipeptide. - After removal of the protecting group, another
protected and activated amino acid can be added
to the N-terminus. - The cycle is repeated until the desired
polypeptide has been synthesized. - It can then be removed from the solid support by
treatment with strong acid. (This also removes
the protecting groups on the amino acid
sidechains that prevent unwanted reactions.)
68
(No Transcript)
69
(No Transcript)
70
(No Transcript)
71
(No Transcript)
72
(No Transcript)
73
Natural polypeptide synthesis
- Performed on the ribosome, a large aggregate of
RNA and protein in effect, "solid phase". - Protecting groups are not necessary, as the
sequence is determined by genetic programming. - The C-terminus is activated by coupling to a
nucleic acid (tRNA). - This tRNA serves both as the activating group and
the connection to the ribosome. The polypeptide
is synthesized starting from the N-terminal amino
acid.
74
(No Transcript)
75
(No Transcript)
76
(No Transcript)
77
(No Transcript)
78
(No Transcript)
79
(No Transcript)
80
(No Transcript)
81
Protein Purification
- In an organism, any one protein is present as a
very small percentage of the total biomolecules.
- In order to characterize a protein fully, it is
necessary to purify it. - There are many possible strategies that one could
use to purify a protein. - In general, the more that you know about the
protein's properties, the better strategy you
could design to purify it. - A typical protein purification strategy will be
comprised of several stages, each one taking
advantage of different characteristics of the
protein.
82
Modulating Solubility
- 1) Precipitation at the pI
- A protein's average net charge at its isoelectric
point is 0. Above or below this pH, the protein
molecules are negatively or positively charged,
respectively, causing them to repel each other.
83
Modulating Solubility
- 2) Salting in
- Many proteins are poorly soluble in pure water,
but are much more soluble in salt-containing
solutions. Thus, lowering ionic strength can be
used to precipitate certain proteins.
84
Solubility of lactoglobin as a function of pH at
several NaCl concentrations.
85
Modulating Solubility
- 3) Salting out
- At very high concentrations (gt1 M) of certain
salts, proteins solubility is reduced due to a
competition with the protein for interaction with
water molecules.
86
Solubility of hemoglobin at its pI as a function
of ionic strength and ion type
87
Ammonium sulfate precipitation
- Precipitation with ammonium sulfate is a common
first step in protein purification. - Because proteins precipitate at different
concentrations of salt, one can perform a first
"cut" with a concentration that will leave the
desired protein soluble, remove the precipitate,
and then add sufficient salt to precipitate the
desired protein.
88
Solubilities of several proteins in ammonium
sulfate solutions.
89
Ammonium sulfate precipitation
- Example
- Your protein will remain soluble at 30 (w/v)
ammonium sulfate, but precipitates at 40
ammonium sulfate. - You slowly add the salt to your protein extract
until it reaches a concentration of 30, allow
precipitation to occur, and then centrifuge the
solution to remove the precipitate. - You take the supernatant, and add ammonium
sulfate until it reaches a concentration of 40,
allow precipitation to occur, and then centrifuge
the solution to remove the precipitate. - You dissolve the precipitate in a buffer and
dialyze it to remove the salt.
90
Dialysis
- The protein is put in a bag of cellulose
membranes having small pores of controlled size.
- Proteins bigger than the pores are retained,
while smaller molecules may diffuse out. - As the volume of the buffer surrounding the bag
is many times (100-1000x) the volume within the
bag, the smaller molecules can be effectively
removed after several changes of the outer buffer.
91
Dialysis
92
Gel filtration chromatography(a.k.a. "molecular
sieve", "size exclusion")
- The protein is applied to the top of a column
consisting of porous beads made of a hydrated
material, such as agarose, dextran, or
polyacrylamide. - The pores of the beads are of a controlled size
and this regulates which proteins can enter the
beads. - The larger proteins will be excluded from the
beads and will flow through the column faster
than the smaller molecules, which experience a
much larger volume. - In practice, there exists a range of pore sizes,
and proteins are separated by their sizes (and
shapes) the largest are eluted first, and the
smallest elute last.
93
Gel filtration chromatography
94
Gel filtration chromatography
- Vbed Vbeads Vvoid
- The void volume is the volume surrounding the
beads. - The bed volume is the total volume of the column.
- Vvoid is typically about Vbed /3
- A protein can be characterized by its elution
volume (Velution), which is the volume of solvent
required to elute it from the column. The
relative elution volume ( Velution / Vvoid) is a
quantity independent of the particular column
used. - By standardizing the column with proteins of
known size, one can use this technique to
estimate molecular weight (assuming that the
shape is close to that of the standards more or
less spherical).
95
(No Transcript)
96
Commonly Used Gel Filtration Materials
97
Ion exchange chromatography
- This technique uses the charge on a protein to
separate it from other proteins. In the process
of ion exchange, ions in solution replace ions
that are electrostatically bound to an inert
support carrying groups with the opposite charge. - Cation exchangers bear negatively charged groups.
- Anion exchangers bear positively charged groups.
- Polyelectrolytes, such as proteins, can bind to
either cation or anion exchangers, depending on
their net charge (i.e. depending on the pH).
98
Ion exchange chromatography
- In most cases, a column is prepared with the ion
exchanger, which is then equilibrated with the
same buffer used to dissolve the protein. The
exchanger and pH are chosen such that the protein
will bind relatively tightly to the ion
exchanger. - The protein solution is loaded to the top and the
column is washed with the buffer, removing all
proteins with the opposite charge or low charge. - The protein can be eluted from the column either
by changing the pH or by increasing the salt
concentration, which shields the charges and thus
decreases their attraction. Elution is most often
carried out by applying a salt gradient to the
column.
99
(No Transcript)
100
Ion exchange chromatographyusing stepwise
elution.
101
Device for generating a linear concentration
gradient.
102
Molecular formulas of cellulose-based ion
exchangers.
103
Some common ion exchangers.
104
Affinity chromatography
- This technique takes advantage of the fact that
many proteins specifically bind other molecules
as part of their function. One can use this
information to construct a column containing the
ligand covalently attached to a matrix. - Upon passing the protein solution through such a
column, only the proteins that can bind the
ligand will be retained on the column. - Then the conditions can be adjusted to effect
release from the ligand. Often this can be done
simply by eluting with the soluble version of the
ligand.
105
(No Transcript)
106
Covalent linking of ligand to agarose
107
Derivatization of epoxy-activated agarose.
108
Example purification of staphylococcal nuclease
by affinity chromatography on bisphosphothymidine-
linked agarose
109
Preparative vs. Analytical
- Preparative methods used to purify proteins
- Able to handle large amounts of protein at once
- The chromatographic techniques just discussed are
all preparative. - Analytical methods used to analyze proteins
- Usually deal with small amounts of protein
- Some preparative techniques can also have
analytical formats.
110
Analytical techniques
- Separation based on
- Mass
- Charge
- Shape
- Different combinations of the above
111
Density Gradients
- An equilibrium sedimentation experiment can be
set up with linear gradients of sucrose or
glycerol. - The protein is loaded on top, and centrifugation
is started. - The advantage of this technique is that the
gradients tend to be rather stable and allow one
to remove the protein of interest from them. The
disadvantage is that they are not so accurate for
determination of S. - Often such gradients are run with molecular
weight standards of known molecular weights to
allow estimation of molecular weight. The
sedimentation coefficient of a protein is
approximately proportional to the 2/3 power of
its molecular weight. Note that this only holds
true for proteins with a globular shape (i.e.
quasi-spherical).Sunknown/Sstandard
(Munknown/Mstandard)2/3
112
Zonal ultracentrifugation
113
Zonal ultracentrifugation
114
Gel electrophoresis
- General technique to analyze mixtures of
proteins, and for limited purification of
proteins. - The protein is driven through a viscous solvent
by an applied electric field (E), due to the
charge of the protein (z) v Ez/f (f the
protein's frictional coefficient) - A proteins electrophoretic mobility (µ) is
defined µ v/E - Typically this is performed in the presence of a
gel support, such as polyacrylamide - prevents convection currents
- enhances separation by serving as a molecular
sieve
115
Polymerization of acrylamide and
N,N-methylenebisacrylamide
116
Native gel electrophoresis
- The native protein migrates through the gel
according to the charge of the protein at the pH
of the buffer system. - Useful for getting relatively pure protein, but
in small amounts. - Not used very often.
117
SDS-polyacrylamide gel electrophoresis(SDS-PAGE)
- Most common form of PAGE, but not useful for
purification of proteins in their native
conformation. - Proteins are solubilized with the detergent SDS
(sodium dodecyl sulfate) - binds polypeptide at a ratio of 1 SDS per 2
amino acid residues - denatures protein ? converts to roughly rod-like
shape - protein has a negative charge roughly
proportional to its mass - The additional negative charge is much greater
than the protein's intrinsic charge, which can
usually be ignored.
118
SDS-polyacrylamide gel electrophoresis(SDS-PAGE)
- As charge/mass ratio is almost constant and the
molecular shapes are all similar, separation is
on the basis of size. - Smaller polypeptides migrate faster and larger
ones migrate slower, due to the gel filtration
effect. - There is an empirical relationship between
mobility and molecular weight µ ? 1/log Mr - Average pore size of the gel can be controlled by
varying the concentration of acrylamide before
initiating polymerization. Higher percentage
polyacrylamide gels (smaller pore sizes) will
result in better resolution of smaller
polypeptides.
119
(No Transcript)
120
slab gel electrophoresis
121
(No Transcript)
122
(No Transcript)
123
(No Transcript)
124
Isoelectric focusing
- A pH gradient is set up by electrophoresing
polyampholytes (300-600 Da oligomers bearing
amino and carboxylate groups in varying rations)
in a gel tube. - The more basic ones (cationic) will accumulate
near the cathode and the more acidic (anionic)
will accumulate near the anode, thereby
establishing a continuous pH gradient. - When a protein is applied to the gel, it will
migrate toward the anode if it is negatively
charged or toward the cathode if positively
charged, until it reaches the region
corresponding to its pI, where it will stop. - Proteins are often denatured in 6M urea, which
does not change the protein's charge (unlike SDS).
125
General formula of the ampholytes used in
isoelectric focusing.
126
Example
- A protein with a pI of 5.2 is introduced near the
cathode end, which has pH of 9.5 (in this gel). - It is then negatively charged and will migrate
toward the anode. - As it migrates in this direction, the pH will
steadily decrease, and the amount of negative
charge carried by the protein will also decrease,
as more and more ionizable groups become
protonated. - Thus it will slow down, until it hits the region
where pH 5.2, where it will experience no
further force. If it diffuses in either
direction, it will pick up charge and the
electric field will force it back. - This results in the protein being concentrated
(focused) in a very narrow region of the gel.
127
(No Transcript)
128
(No Transcript)
129
2-dimensional gels
- This is a powerful technique that combines
isoelectric focusing with SDS-PAGE. - Proteins are separated in the first dimension by
isoelectric focusing (IEF). - Then this tube is attached to the side of an
SDS-polyacrylamide gel. SDS-PAGE provides the
second dimension. - Each protein migrates to a semi-unique spot
according to its pI and molecular weight (MW).
130
(No Transcript)
131
(No Transcript)
132
Two-dimensional (2D) gel electrophoresis
133
Protein Purification Practical Aspects
- All cells contain proteases enzymes that
catalyze hydrolysis of peptide bonds. - Upon breaking cells, these are released into the
extract, where they can degrade the protein you
want to purify. - In order to inhibit proteolysis and denaturation,
protein purification is usually carried out in
the cold (on ice or in a cold room) in the
presence of protease inhibitors (small molecules
that inhibit specific proteases).
134
Protein Purification Practical Aspects
- You often have to choose your source of material
carefully - unicellular organism
- organ of a metazoan (or plant)?
- Where is the enzyme located?
- In cytosol?
- Within membrane-bound organelle?
- Secreted?
- You can use differential centrifugation to do a
(crude) separation of various cellular
compartments to get a starting point. - If the protein is in a specific organelle,
density gradient centrifugation is usually used
to purify the organelle first.
135
(No Transcript)
136
(No Transcript)
137
(No Transcript)
138
(No Transcript)
139
Purification table
Activity definition of 1 unit (U) will vary,
depending upon enzyme Specific activity measure
of enzymes purity activity/protein Yield of a
step Units retained after that step/Units
input(Measure of how well enzyme activity is
preserved by the step.) Purification factor
specific activity after the step/before(Measure
of how effective the step is.)
140
Purification table
- Yield and purification factor can be expressed
- for the individual step OR
- as cumulative yield or purification factor,
taking into account all steps up to to that
point - You need to be careful to distinguish between
them.
141
Example Purification of Rat Liver Glucokinase
142
Modern tricks
- Genetic engineering assists protein purification
- In the past, one often had to purify a protein in
order to obtain enough sequence information to
search for the gene. - Nowadays, one often has a gene encoding a protein
long before the protein has been purified. - In either case, "engineering" the gene can ease
protein purification.
143
Modern tricks
- One can use strong promoters (DNA elements that
direct RNA polymerase to transcribe the gene) to
overproduce the protein, either in a homologous
system, or in a very different system. This
increases the specific activity of your starting
material. - One can attach sequence elements to the gene,
such that the protein will have additional
polypeptide segments that can be used as "tags"
for affinity purification. - Example The sequence H-H-H-H-H-H (hexahistidine)
is capable of binding Ni2 ions. Attachment of
the "His6-tag" to a protein allows it to bind to
a nickel-nitrilotriacetic acid (NTA-Ni2) column
and be eluted with imidazole.
144
Example Engineering removable affinity tag
- Clone gene into plasmid to express in bacteria
- Fuse protein to intein and chitin-binding domain
(CBD) - Makes chimeric protein
- Pass through chitin column and wash off all other
proteins. - DTT allows intein to cleave link between itself
and protein. - Protein elutes, leaving intein-CBD on the column.