Microcrystals PowerPoint PPT Presentation
Title: Microcrystals
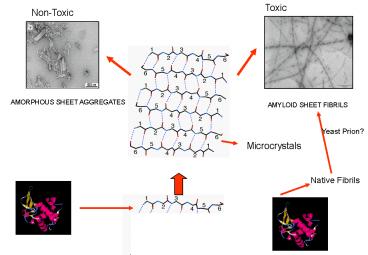
1
Yeast Prion?
Microcrystals
Native Fibrils
2
Yeast Prion?
Microcrystals
SHEET AGGREGATES
Native Fibrils
3
Tango A software to predict aggregation prone
regions in proteins (http//tango.embl.de/)
..\..\Documents and Settings\user\Desktop\tangore
d2.gif Uses the Fold-X force field and AGADIR
(http//www.embl-heidelberg.de/Services
http//model-x.embl.de/) Assumes 100 burial of
the amino acid sequence Considers competing
conformations (a-helix and b-turn) Takes into
account pH, TFE, temperature, Ionic strenght,
Protein Stability and concentration
Fernandez-Escamilla, A. M., F. Rousseau, et al.
(2004). "Prediction of sequence-dependent and
mutational effects on the aggregation of peptides
and proteins." Nat Biotechnol 22(10) 1302-6.
4
Tango Prediction of Aggregation Prone Regions
Acp
a-Syn
Experimentally Detected
B4A4
Tau PHF43
Fernandez-Escamilla, A. M., F. Rousseau, et al.
(2004). Nat Biotechnol 22(10) 1302-6.
5
Predictions for 176 peptides
predicted under 1 93
hit rate 95
false negatives 5
false negatives 5
predicted between 1-5 10
predicted positive 4
positive 40
predicted negative 6
negative 60
predicted above 5 73
hit rate 82
false positives 13
false positives 18
6
Human Lysozyme
ddG Agg ___________________ WT
0 f57i 3.72 0.6 i56t 3.10 0.3 d67h 3.88 0.8 w64r 3
.40 0.4
7
Transthyretin
ddG
Aggr ________________________ WT
0.7 d18g. 24.1 42.8 p24s. 8.1 24.5 t119m -2.7
0.0 v20i 0.60 1.7 v122i -1.7 0.1 a45t. 0.09 0.9 d
18g 9.11 42.8 g53e 15.2 40.9 l12p 10.8 40.9 l55p 1
0.7 40.8 l111m 1.4 5.7 v30g 12.8 36.6 v30m 4.35 1
8.4
8
Yeast Prion?
Microcrystals
Native Fibrils
9
Sequence determinants of amyloid fibril formation
and toxicity
Luis Serrano European Molecular Biology
Laboratory (Heidelberg, Germany)
10
Amyloidogenic polypeptides
- Non-pathogenic proteins
PI3-SH3
Acetylphosphatase
Myoglobine
Guijarro, J. L. et al. PNAS 95, 4224-8 (1998)
Chiti F. et al. PNAS 96, 3590-4 (1999)
Fandrich, M. Dobson, C. Nature 410, 165-6 (2001)
pH 5.5 25 v/v TFE
pH 2.0
pH 9.0 65 C
Taken from Dobon and co-workers
11
The Amyloid Stretch Hypothesis
Short sequences are enough to recruit Proteins
to the Dark Side
a-spectrin SH3
21DIDLHLGD28
12
Short aa stretches are responsible for fibril
formation.
SPC-SH3 MDETGKELVLALYDYQEKSPREVTMKKGDILTLLNS------
---------TNKDWWKVEVNDRQGFVPAAYVKKLD PI3-SH3
-MAEGYQYRALYDYKKEREEDIDLHLGDILTVNKGSLVALGFSDGQEAR
PEEIGWLNGYNETTGERGDFPGTYVEYIGRKKISP
PI3-SH3
amyloid fibrils at acidic pH
21DIDLHLGD28
TOXIC
Dobson, Aviles Serrano Groups. PNAS 2004
13
1-SH GGSTVIIE------- 2-SH
GGSTVVIT----- 3-SH
GGSTVIKT------ - AB-SH
GGKLVFFA---- SH-1
-----STVIIEGG SH-2
-----STVVTGG SH-3
-----STVIKTGG AmyBergerac
-------STVIIE--------
Esteras et al., PNAS (2005).
14
Isolation of the Amyloid Core
STVIIEGG-------------
15
Conclusions(I)
- Demonstration of the amyloid stretch hypothesis
Short amyloidogenic stretches can trigger
amyloid fibril formation by a
non-amyloidogenic protein. ?
Both de novo and natural sequences.
- In order to be amyloidogenic a protein must
carry an appropiate - amyloid sequence (Sequence determinant) that
must become - locally unfolded (Structural determinant) to
initiate the process.
- Point mutations in the amyloidogenic stretch
designed with the amyloid - pattern can modulate the amyloid behaviour of
a protein.
- The amyloid insertion is incorporated into the
amyloid core of the fibril and - can trigger the incorporation of some other
regions of the protein as well.
Esteras et al., PNAS (2005).
16
Peptide-based model system for amyloidogenesis
- Assumption
- Propagation and stacking of the pre-formed
b-sheets would result in the final assembly of
amyloid fibrils - Design Method
- Protein design algorithm - PERLA
- Modeled Amino Acids
- Positions 1 6 KETS
- Positions 2 5 KETSLVIYW
- Positions 3 4 LVIYW
- Backbone Template
- 6-Stranded anti-parallel b-sheet with six
residues per strand
López de la Paz, M. et al. Proc. Natl. Acad. Sci.
USA 99, 16052-7 (2002)
17
Designed favourable sequences aggregate as
b-sheets There is no obligatory correlation
between b-sheet formation and amyloid formation.
Fibrils
The predicted sequence stability is No. 1 gt 3 gt 6
gt 7 gt 8 gt 9 gt 5 gt 4.
18
STVIIE
12.1 Å
10.0 Å
10.0 Å
19
Positional scanning mutagenesis on STVIIE
1 XThrValIleIleGlu 2 Ser
XValIleIleGlu 3 SerThrXIleIleGlu 4
SerThrValXIleGlu 5 SerThrValIleXGlu
6 SerThrValIleIle X
X natural amino acids, except Cys net charge
1 pH 2.6 (amyloid fibril formation in vitro)
7.4 (amyloidoses- in vivo)
STVIIE pH 2.6 (net charge 1)
López de la Paz Serrano PNAS 2004
20
Positional scanning EM fibril detection
net charge 1 2
3 4 5 6 Ala (1)
Met (1) Phe
(1) Val (1)
Ile (1)
Leu (1)
Ser (1) Thr (1)
Tyr (1)
Trp (1)
Asn (1) Gln (1)
Glu0 (1)
Glu (1)
Asp0 (1) Asp (1)
Gly (1)
Pro (1) Lys (1)
Arg (1)
His (1) His0 (1)
21
Sequence-based detection of amyloidogenic protein
regions
López de la Paz Serrano PNAS 2004
22
Toxic
Non-Toxic
Amyscan, Structure-Based Rational Design
Tango
23
Sequence-based detection of amyloidogenic protein
regions
López de la Paz Serrano PNAS 2004
24
Sequence scanning of amyloid proteins
Beta-2-microglobulin (b2m)
Thr
Tyr
Predicted (pH 2.6 7.4) Experimental Site
63-68, Identity Region 61-70 kdws_FYLLYY_teft WSF
YLLYYTE Site 64-69, Identity dwsf_YLLYYT_eftp
Tyr
Leu
Leu
Tyr
Phe
- McParland, V. J., et al. Nat Struct Biol 9,
326-31 (2002) - Jones, S. et al. J Mol Biol 325, 249-57 (2003)
25
Pattern Validation Sequence Scanning of Amyloid
Proteins (66 success)
Gelsolin LMSLFG - Human Insulin QLENYC CSLYQ
- NYCNFV - RGFFYT - Human PrP
N- KGENFT - MLVLFV NQNNFV DCVNIT Huma
n PrP (C-terminal part) SMVLFS VILLIS
ISFLIF SFLIFL LIFLIV Human
IAPP SSNNFG - Apolipoprotein A1 VTQEFW - LAVLF
L VTSTFS ALEEYT Bacterial
CSP VSFEIV - Acylphosphatase EYSNFS - Protein
G GEWTYD ?-synuclein EGVLYV PI3-SH3 TYVEYI
YVEYIG Ig Kappa IgI YLNWYQ Ig Kappa
IgII GVNYFL Ig Kappa IgIII YTFTIS - Sup35
(Yeast PrP) KNFNYNI Tau KVQIIN Cystatin
C-1 AGVNYF Cystatin C-2 GVNYFL Cystatin
C-3 SFQIYA Hen lysozyme FESNFN Sup35 (Yeast
PrP) NQQNQY YYQNYQ
3mg/ml, 1 Month incubation. In some of the
negative cases fibers appeared after 3 months
26
Conclusions(II)
- Short amyloidogenic streches can make a protein
become amyloidogenic.
- Using a variety of methods we could identify
some of these amyloidogenic - Segments, whichh could be used for drug screening
- Amyloidoses can be triggered by
- Point mutations that destabilize a folded state
- Point mutations that create an amyloidogenic
sequence - Postranslational modifications that overcome
natural amyloid breakers (i.e AB1-42) - Binding of molecules that trigger conformational
changes (i.e Cu) - Problems in the cell maintenance machinery
(proteosome, chaperones) - Etc..
27
The Amyloid Stretch Hypothesis
Amyloid sequences and toxicity
Is there a sequence dependent on toxicity?.
Pastor, M et al In preparation (collaboration
with Carlos Dottis group, Milan)
28
Table 1.- Amyloidogenic stretches. Set of
amyloidogenic stretches selected for this study
and obtained by protein sequence scanning for
6-residue fragments matching the amyloid pattern
(De la Paz et al., PNAS. 2004).
29
Morphology of mature fibrils.
a
d
c
b
h
g
f
e
Electron micrographs showing the morphology of
matured fibrils formed by the amyloid stretches
selected (a) Tau590-595 , (b) Ab16-21 , (c)
PrP178-183 , (d) PrP244-249 , (e) PrP245-250 ,
(f) CysC98-103 , (g) ApoA18-13 and (h) STVIIE.
30
Toxicity of mature fibrils and monomeric
hexapeptides
a
d
c
b
h
g
f
e
a) PC12 cells viability of the hexapeptides
selected in different states of amyloid fibril
formation. Black bars refer to monomeric peptide
and grey bars to their corresponding mature
fibrils
31
Cytotoxicity of Tau590-595 species in PC12 cells.
a
Mature Fibrils Sonicated Fibrils
Re-polymerized fibrils
b
c
d
a) Cytotoxic effect of monomers, mature fibrils,
protofibrils trapped by monitoring the
self-assembly reaction course (I) and by
sonication (II) of mature fibrils b-d) Electron
micrographs showing (b) mature fibrils of
Tau590-595 (c) Tau590-595 toxic protofibrils
generated by sonication and (d) Tau590-595 mature
fibrils obtained after the incubation of toxic
protofibrils obtained by sonication.
32
Monomers, fibrils and species generated by
ultrasonication of the amyloidogenic stretches
a
TAU AB
Prion CT3 Prion CT4
Cystatin
a) PC12 cells viability of protofibrils generated
by ultrasonication and dilution of their
corresponding mature fibrils at different
concentrations. Black bars refer to 50 mM, grey
bars to10mM and white bars to 5 mM. b-f) Electron
micrographs showing the morphology of the toxic
prefibrillar aggregates generated as it has been
described below of amyloid stretches of tau (b),
Ab (c), prion Ct3 (d), prion Ct4 (e) and cystatin
C (f).
33
Cytotoxicity of Ab1-42 protofibrils
AB1-42 Fibrils Ab1-42 protofibrils
generated by
ultrasonication
c
Electron micrographs of a) Ab1-42 fibrils and b)
Ab1-42 protofibrils generated by ultrasonication
of mature fibrils. c) Pathogenic effect in PC12
cells of monomers, fibrils and protofibrils of
Ab1-42 trapped by monitoring the self-assembly
reaction course (I) and by sonication of mature
fibrils (II) and protofibrils of Ab16-21.
Protofibrils of amyloidogenic natural protein
show similar toxicity, regardless the strategy
used to obtained them. Moreover, these
protofibrils provoke similar reduction in PC12
cell viability to protofibrils of the
amyloidogenic stretch of this protein.
34
Morphology of non-toxic aggregates
Electron micrographs showing the morphology of
species generated by ultrasonication and dilution
of mature fibrils of (a) prion PrP178-183 (b)
ApoA18-13 and (c) STVIIE. Although mature fibrils
were sonicated and diluted under a broad of range
of conditions, the species obtained did not
affect to PC12 viability.
35
Toxicity of STVIIE in different states of amyloid
formation
Sonication of mature fibrils under a broad range
of conditions failed to generate protofibrils.
The species generated by sonication (brick-like
morphology) were toxic. Protofibrils obtained by
monitoring the amyloid formation reaction are
cytotoxic for PC12 cells.
36
Toxicity is independent of sequence but related
to structure of the aggregates
NON-TOXIC
TOXIC
37
Acknowledgments
Amyloid subgroup Sandra
Esteras
Mayte Pastor Manuela Lopez de la Paz
Niko Kuemmere
EMBL, HD Emmanuel Lacroix (PERLA) Kenneth Goldie
(cryo-EM) Andreas Hoenger (shadowing) Salvador
Ventura (a-spectrin vs. PI3-SH3) Groups Christoph
er C. Dobson Carlos Dotti Marcos Milan Louise
Serpell Xavier Aviles
Financed by EC grants Apopis and
HPRN-CT-2002-00241
38
The Amyloid Stretch Hypothesis
Amyloid sequences and toxicity
Fibril toxicity
Pastor, M et al In preparation (collaboration
with Carlos Dottis group, Milan)
39
Plasma membrane interaction and internalization
of protofibrils in PC12 cells
a
a) Citotoxicity of monomeric species, fibrils and
protofibril of fluorescein-Tau590-595 and
fluorescein-Ab16-20. Protofibrils of
fluorescein-labeled peptides are less toxic that
protofibrils of Tau590-595 and Ab peptides. b-c)
Confocal microscopy images showing protofibrils
of tau-fluorescein (b) in contact with plasma
membrane and cytoplasm and (c) in the cell
nucleus. Cells were incubated for 4 h in the
presence of the protofibrils. Arrowheads
indicate aggregates within the cell.
40
Toxicty is chirality Indepent
Toxicity of monomers, mature fibrils and
protofibrils of D-Tau peptide in PC12 cells.
Protofibrils of Tau590-595 and its corresponding
D-enantiomer show similar cell reduction,
suggesting that does not exist a chiral
recognition between toxic species and their
target.
Apoptosis induced by protofibrils on PC12 cells.
Apoptosis range of untreated cells, cells
incubated with monomers, mature fibrils and
protofibrils of Tau590-595 during 4 hours.
Incubation of protofibrils of D- Tau590-595
provoke a similar percentage of apoptosis to
protofibrils of L-Tau590-595.
41
Protofibril aggregates localize in synapses, are
internalized and disorganize the actin network
42
(No Transcript)
43
Tango A software to predict aggregation prone
regions in proteins (http//tango.embl.de/)
Validated with 1000 peptide analysis of
aggregation
ADA2H
Fernandez-Escamilla, A. M., F. Rousseau, et al.
(2004). Nat Biotechnol 22(10) 1302-6.
44
(No Transcript)
45
The Amyloid Stretch Hypothesis
Recruiting Proteins to the Dark Side and
Designing Inhibitors to Bring them Back
Short amino acid stretches bearing a highly
amyloidogenic motif could be used to screen for
inhibitors.
A. Esteras-Chopo, M. Lopez de la Paz and L.
Serrano In preparation
46
Short Sequences as Targets for Amyloid Inhibition
Screen for inhibitors of the amyloid region
Validate leads against amyloid protein
47
Deconvolution of a positional scanning D-library
- LIBRARY DESIGN
- Ac-GGOXXXXXGcG-NH2
- Ac-GGXOXXXXGcG-NH2 O one of the 20
D-aa - Ac-GGXXOXXXGcG-NH2 X close to
equimolar mixture of - Ac-GGXXXOXXGcG-NH2 D-aa
except D-Cys. - Ac-GGXXXXOXGcG-NH2 120 peptide
mixtures ,each containing - Ac-GGXXXXXOGcG-NH2 (19)5 individual
undecapeptides.
TARGET De novo designed sequence STVIIE (1)
TOOLS CD?-sheet population after inhibition /
?-sheet population stock.
EMEvaluation changes in amount and morphology of
fibrillar material.
48
Short Sequences as Targets for Amyloid Inhibition
49
Result of the Deconvolution of a D-Peptide Library
Better inhibition gt gt gt -
SELECTION CRITERIA avoid charged aminoacids
aminoacids ranked with or
Esteras-Chopo et al. In preparation.
50
Design and Testing of Defined D- Sequences
Assumption The combination of the best
aminoacids at every position
will generate the best possible inhibitors.
96 defined sequences 32 for
testing against mature and sonicated fibrils
A) Set of lead sequences inhibition of 1-SH
INHIBITOR A
MATURE STVIIE
STVIIE
1WEEK
MATURE STVIIE-SH
Esteras-Chopo et al. In preparation.
51
B) Set of Empirical rules for the Design of
D-inhibitors
A? stretch (KLVFFA)
SA? TAU stretch (KVQIIN
) STAU
A? STRETCH
TAU STRETCH
Esteras-Chopo et al. In preparation.
52
SCREEN FOR LEADS AGAINST THE AMYLOID STRETCH
VALIDATE LEADS AGAINST AMYLOID PROTEIN
AMYLOID PATTERN
hjlope
AMYLOID STRETCH
Esteras-Chopo et al. In preparation.
53
Figure 7 of supplementary material.- Apoptosis
induced by protofibrils on PC12 cells. Apoptosis
range of untreated cells, cells incubated with
monomers, mature fibrils and protofibrils of
Tau590-595 during 4 hours. Incubation of
protofibrils of D- Tau590-595 provoke a similar
percentage of apoptosis to protofibrils of
L-Tau590-595.
54
Electron micrographs showing the morphology of
protofibrils of Ab (1-42).
a) Protofibrils generated by sonication of mature
fibrils and b) protofibrils caught by following
the kinetic of fibril formation. Both of them
shown similar morphology, shorter and thinner
than mature fibrils