The Rat Genome Database: PowerPoint PPT Presentation
1 / 1
Title: The Rat Genome Database:
1
The Rat Genome Database Automated pipelines
facilitate rapid and reliable updates of genomic
data
Jennifer Smith, Dawei Li, George Kowalski, Mary
Shimoyama, Andrew Patzer, Victoria Petri, Stan
Laulederkind, Rajni Nigam, Jeff De Pons, Melinda
Dwinell, Simon Twigger, Howard Jacob Rat Genome
Database, Human and Molecular Genetics Center,
Medical College of Wisconsin
Abstract
Biological data, like the organisms it describes,
grows and changes on a daily basis. As such, it
is essential that the databases which house such
data employ fast and efficient methods of
updating the information that they store. The
Rat Genome Database (http//rgd.mcw.edu) is a
publicly available resource which warehouses data
on the rat as a model organism for human
physiology and disease as well as rat, mouse and
human comparative genomic data. RGD has recently
implemented a series of pipelines which rapidly
and reliably import data from the Entrez Gene
database at NCBI. Each week, the rat, mouse and
human Entrez Gene pipelines automatically query
the NCBI databases for gene records that have
been modified during that week. These records
are downloaded as XML files, which are parsed and
the relevant data extracted for each record.
Incoming records are matched to corresponding RGD
records by matching on Entrez Gene ID, one
nucleotide sequence ID, and, depending on the
species of the gene, the RGD, MGI, or HGNC or
HPRD ID. If a match is made, the pertinent data
in RGD, including sequence identifiers and
genomic positions, is deleted and subsequently
reloaded from the Entrez Gene record eliminating
the need for time-consuming checks of each
individual piece of data and thus substantially
improving performance. Entrez Gene records with
no match in RGD are loaded as new genes. Log
files track which genes were updated and which
incoming records were rejected. A conflict log
allows curators to manually review loading
problems. Frequent incremental updates have
reduced the previously labor-intensive process of
manually reviewing conflicts from an average two
weeks to a few hours. In addition, with
multiple pipelines forming a network of data
checks, RGD is substantially improving both the
quality and quantity of data we supply to our
users.
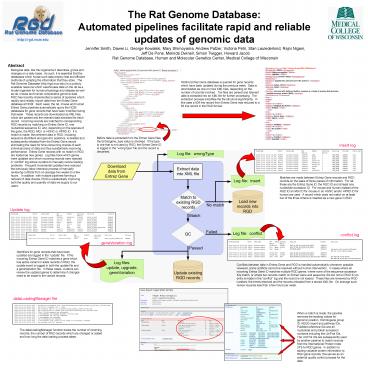
NCBIs Entrez Gene database is queried for gene
records which have been updated during the
previous week. Data is downloaded as one or more
XML files, depending on the number of records
involved. The files are parsed and relevant data
is extracted into an XML file for further
processing. The extraction process simplifies
the file structure significantly. In this case a
504 line record from Entrez Gene was reduced to a
63 line record in the RGD format.
Before data is extracted from the Entrez Gene
files, the Entrezgene_type value is checked. If
the type is one that is not used by RGD, the
Entrez Gene ID is logged in the wrongType file
and the record is discarded.
insert log
Matches are made between Entrez Gene records and
RGD records on the basis of three pieces of
information. For rat these are the Entrez Gene
ID, the RGD ID and at least one nucleotide
accession ID. For mouse and human instead of the
RGD ID an MGI ID for mouse or an HGNC and/or HPRD
ID for human are used. A record which does not
match on at least two of the three criteria is
inserted as a new gene in RGD.
conflict log
Identifiers for gene records that have been
updated are logged in the update file. If the
incoming Entrez Gene ID matches a gene which has
splice variant or allele records in RGD, the
update event is logged in both the update file
and a geneVariation file. In these cases,
curators can review the updated genes to
determine if changes need to be made to the
variant records.
Conflicts between data in Entrez Gene and RGD is
handled automatically whenever possible. However,
some conflicts cannot be resolved without human
intervention. In cases where an incoming Entrez
Gene ID matches multiple RGD genes, where none of
the sequence accession IDs match, or where two
records match on Entrez Gene and sequence IDs but
not on RGD ID an entry is made in the conflict
log and the record is not loaded. These files
are reviewed by RGD curators, the errors resolved
and the records reloaded from a stored XML file.
On average such review requires less than a few
hours per week.
dataLoadingManager file
When a match is made, the pipeline removes the
existing values for genomic position, Homologene
group ID, KEGG report and pathway IDs, PubMed
reference IDs and all nucleotide and protein
accession numbers including the UniProt IDs. The
UniProt IDs are subsequently used by another
pipeline to match records from the International
Protein Index (IPI) to RGD genes. In addition to
adding valuable protein information to RGD gene
records, this serves as an external quality
control process for the data.
The dataLoadingManager function tracks the number
of incoming records, the number of RGD records
which are changed or added and how long the data
loading process takes.