Comparative Annotation of Viral Genomes with Non-Conserved Gene Structure PowerPoint PPT Presentation
Title: Comparative Annotation of Viral Genomes with Non-Conserved Gene Structure
1
Comparative Annotation of Viral Genomes with
Non-Conserved Gene Structure
Saskia de Groot and Jotun Hein Department of
Statistics, University of Oxford
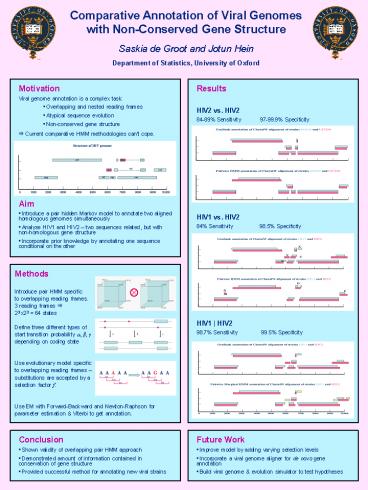
- Motivation
- Viral genome annotation is a complex task
- Overlapping and nested reading frames
- Atypical sequence evolution
- Non-conserved gene structure
Results HIV2 vs. HIV2 84-89 Sensitivity
97-99.9 Specificity HIV1 vs.
HIV2 84 Sensitivity 98.5
Specificity HIV1 HIV2 98.7
Sensitivity 99.5 Specificity
? Current comparative HMM methodologies cant
cope.
- Aim
- Introduce a pair hidden Markov model to annotate
two aligned homologous genomes simultaneously - Analyse HIV1 and HIV2 two sequences related,
but with non-homologous gene structure - Incorporate prior knowledge by annotating one
sequence conditional on the other
Methods Introduce pair HMM specific to
overlapping reading frames. 3 reading frames
? 23 x23 64 states Define three different
types of start transition probability a, ß,
? depending on coding state Use evolutionary
model specific to overlapping reading frames
substitutions are accepted by a selection
factor f. Use EM with Forward-Backward and
Newton-Raphson for parameter estimation
Viterbi to get annotation.
- Conclusion
- Shown validity of overlapping pair HMM approach
- Demonstrated amount of information contained in
conservation of gene structure - Provided successful method for annotating new
viral strains
- Future Work
- Improve model by adding varying selection levels
- Incorporate a viral genome aligner for de novo
gene annotation - Build viral genome evolution simulator to test
hypotheses
PowerShow.com is a leading presentation sharing website. It has millions of presentations already uploaded and available with 1,000s more being uploaded by its users every day. Whatever your area of interest, here you’ll be able to find and view presentations you’ll love and possibly download. And, best of all, it is completely free and easy to use.
You might even have a presentation you’d like to share with others. If so, just upload it to PowerShow.com. We’ll convert it to an HTML5 slideshow that includes all the media types you’ve already added: audio, video, music, pictures, animations and transition effects. Then you can share it with your target audience as well as PowerShow.com’s millions of monthly visitors. And, again, it’s all free.
About the Developers
PowerShow.com is brought to you by CrystalGraphics, the award-winning developer and market-leading publisher of rich-media enhancement products for presentations. Our product offerings include millions of PowerPoint templates, diagrams, animated 3D characters and more.