Biological Databases - PowerPoint PPT Presentation
1 / 44
Title:
Biological Databases
Description:
An organized body of persistent data and associated ... honeybee. sea urchin. zebrafish. cow. dog. black poplar. RefSeq Accession Numbers. mRNAs and Proteins ... – PowerPoint PPT presentation
Number of Views:91
Avg rating:3.0/5.0
Title: Biological Databases
1
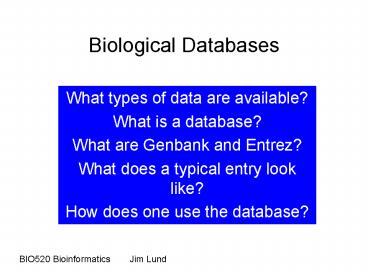
Biological Databases
- What types of data are available?
- What is a database?
- What are Genbank and Entrez?
- What does a typical entry look like?
- How does one use the database?
BIO520 Bioinformatics Jim Lund
2
Biological Data
- Central Dogma-o-centric
- Genomic DNA sequence
- mRNA/cDNA sequence
- Protein sequence
- Protein 3D structure
- Literature (Function)
3
Biological Data
- Genomic DNA sequence (complete)
- mRNA/cDNA sequence
- Gene expression data (NEW)
- Microarrays, SAGE
- Expression catalogs
- Protein sequence
- Protein interaction/complex data (NEW)
- Protein 3D structure
- Literature (Function)
- Organism databases (NEW)
- Annotation and classification projects (NEW)
4
What is a Biological Database?
- An organized body of persistent data and
associated computer software for updating,
querying, and retrieving data records. - Collection of records and files
- Organized for a particular purpose
- The database is separate from the interface and
can have several interfaces. - NCBI Protein can be searched by protein name or
using BLAST (Basic Local Alignment Search Tool).
5
Common database features
- Relational Databases
- Tables
- Relationships between tables
- Version Control
- Consistency enforcement
- Multiauthor/multiuser with security
6
BIO520 Student Database
Table
- 2005
- Name ID Grade
- Amy 123 A
- Joe 456 B
- Sue 789 C
.
Record
Attribute
7
Genbank Entry
LOCUS BC005255 495 bp
mRNA linear PRI 23-JUN-2006 DEFINITION Homo
sapiens insulin, mRNA (cDNA clone IMAGE3950204),
complete cds. ACCESSION BC005255 VERSION
BC005255.1 GI13528923 KEYWORDS MGC. SOURCE
Homo sapiens (human) ORGANISM Homo sapiens
Eukaryota Metazoa Chordata
Craniata Vertebrata Euteleostomi
Mammalia Eutheria Euarchontoglires Primates
Haplorrhini Catarrhini Hominidae
Homo. FEATURES Location/Qualifiers sou
rce 1..495
/organism"Homo sapiens" gene 1..495
/gene"INS"
/db_xref"GeneID3630" CDS 60..392
/gene"INS"
/translation"MALWMRLLPLLALLALWGPDPAAAFVNQHLCGS
HLVEALYLVCG ERGFFYTPKTRREAEDLQVGQVELGGGPGAGSLQPLAL
EGSLQKRGIVEQCCTSICSL
YQLENYCN" ORIGIN 1 agccctccag
gacaggctgc atcagaagag gccatcaagc agatcactgt
ccttctgcca 421 ccgcctcctg caccgagaga
gatggaataa agcccttgaa ccaacaaaaa aaaaaaaaaa
481 aaaaaaaaaa aaaaa //
8
The CORE DDBJ, EMBL, and Genbank
9
Genbank DNA Sequence Database
- Genbank/EMBL/DDBJ Mirror exchange sequence
records. - Primary vs Secondary Databases
- nr (non-redundant database)
- Primary vs secondary records
- Sequence vs inferred property (coding region)
- Format vs content
10
Primary vs. Derivative Databases
- Primary Databases
- Original submissions by experimentalists
- Content controlled by the submitter
- Examples GenBank, SNP, GEO
- Derivative Databases
- Built from primary data
- Content controlled by third party (NCBI)
- Examples Refseq, TPA, RefSNP, UniGene, NCBI
Protein, Structure, Conserved Domain
11
A TraditionalGenBank Record
LOCUS AY182241 1931 bp
mRNA linear PLN 04-MAY-2004 DEFINITION
Malus x domestica (E,E)-alpha-farnesene synthase
(AFS1) mRNA, complete cds. ACCESSION
AY182241 VERSION AY182241.2
GI32265057 KEYWORDS . SOURCE Malus x
domestica (cultivated apple) ORGANISM Malus x
domestica Eukaryota Viridiplantae
Streptophyta Embryophyta Tracheophyta
Spermatophyta Magnoliophyta eudicotyledons
core eudicots rosids eurosids I
Rosales Rosaceae Maloideae Malus. REFERENCE
1 (bases 1 to 1931) AUTHORS Pechous,S.W. and
Whitaker,B.D. TITLE Cloning and functional
expression of an (E,E)-alpha-farnesene
synthase cDNA from peel tissue of apple fruit
JOURNAL Planta 219, 84-94 (2004) REFERENCE 2
(bases 1 to 1931) AUTHORS Pechous,S.W. and
Whitaker,B.D. TITLE Direct Submission
JOURNAL Submitted (18-NOV-2002) PSI-Produce
Quality and Safety Lab, USDA-ARS,
10300 Baltimore Ave. Bldg. 002, Rm. 205,
Beltsville, MD 20705, USA REFERENCE
3 (bases 1 to 1931) AUTHORS Pechous,S.W. and
Whitaker,B.D. TITLE Direct Submission
JOURNAL Submitted (25-JUN-2003) PSI-Produce
Quality and Safety Lab, USDA-ARS,
10300 Baltimore Ave. Bldg. 002, Rm. 205,
Beltsville, MD 20705, USA REMARK
Sequence update by submitter COMMENT On Jun
26, 2003 this sequence version replaced
gi27804758. FEATURES
Location/Qualifiers source 1..1931
/organism"Malus x
domestica" /mol_type"mRNA"
/cultivar"'Law Rome'"
/db_xref"taxon3750"
/tissue_type"peel" gene
1..1931 /gene"AFS1"
CDS 54..1784
/gene"AFS1" /note"terpene
synthase" /codon_start1
/product"(E,E)-alpha-farnesene
synthase" /protein_id"AAO228
48.2" /db_xref"GI32265058"
/translation"MEFRVHLQADNEQKI
FQNQMKPEPEASYLINQRRSANYKPNIWK
NDFLDQSLISKYDGDEYRKLSEKLIEEVKIYISAETMDLVAKLELIDSV
RKLGLANLF EKEIKEALDSIAAIESDNL
GTRDDLYGTALHFKILRQHGYKVSQDIFGRFMDEKGTLE
DFLHKNEDLLYNISLIVRLNNDLGTSAAEQERGDSPSSI
VCYMREVNASEETARKNIK
GMIDNAWKKVNGKCFTTNQVPFLSSFMNNATNMARVAHSLYKDGDGFGDQ
EKGPRTHI LSLLFQPLVN" ORIGIN
1 ttcttgtatc ccaaacatct cgagcttctt
gtacaccaaa ttaggtattc actatggaat 61
tcagagttca cttgcaagct gataatgagc agaaaatttt
tcaaaaccag atgaaacccg 121 aacctgaagc
ctcttacttg attaatcaaa gacggtctgc aaattacaag
ccaaatattt 181 ggaagaacga tttcctagat
caatctctta tcagcaaata cgatggagat gagtatcgga
241 agctgtctga gaagttaata gaagaagtta agatttatat
atctgctgaa acaatggatt //
The Flatfile Format
12
Genbank Entry
LOCUS PCU30791 1234 bp mRNA
PLN 31-MAY-1996 DEFINITION Pneumocystis
carinii carinii form 6 guanine nucleotide binding
protein alpha subunit (pcg1) mRNA, complete
cds. ACCESSION U30791 NID
g1345098 VERSION U30791.1 GI1345098
Unique ID Version Control
13
Content-Taxonomy
SOURCE Pneumocystis carinii f. sp. carinii.
ORGANISM Pneumocystis carinii f. sp. carinii
Eukaryota Fungi Ascomycota
Archiascomycetes Pneumocystidaceae Pneumocystis.
14
Reference
REFERENCE 1 (bases 1 to 1234) AUTHORS
Smulian,A.G., Ryan,M., Staben,C. and Cushion,M.
TITLE Signal transduction in Pneumocystis
carinii characterization of the genes (pcg1)
encoding the alpha subunit of the G protein
(PCG1) of Pneumocystis carinii carinii and
Pneumocystis carinii ratti JOURNAL Infect.
Immun. 64 (3), 691-701 (1996) PUBMED 96186460
- Unique crossreferent
- Can be gt1 reference
15
Features
FEATURES Location/Qualifiers source 1..1234
/organism"Pneumocystis carinii f. sp.
carinii /strain"Form 6 /note"450 kb
chromosome" /db_xref"taxon38081 5'UTR
1..90 gene 91..1155 /gene"pcg1"
Correct?
16
CDS
CDS 91..1155 /gene"pcg1 /note"G-protein
alpha subunit" /codon_start1
/product "guanosine nucleotide binding
protein alpha subunit"
/protein_id"AAC49295.1"
/db_xref"PIDg1345099"
/db_xref"GI1345099"
/translation"MGCCFSATYNQDTLRSKEIE
SYLRQEQEHACHEAKILLLGAGES
.
INFERRED
17
DNA
BASE COUNT 421 a 171 c 195 g 447 t ORIGIN
1 tgaattctaa attttatatt 1201 tattttttta
tgctccagat aaaa //
18
Genbank entries
- Combination of required (LOCUS, SOURCE) and
optional fields. - The entry is hierarchical, some fields contain
subfields. REFERENCE-gtAUTHORS - Some fields can appear multiple times (REFERENCE,
/gene) - Some fields are numerical, other are text. Some
fields contain free text, others use a controlled
vocabulary or an database ID.
19
Other Genbank output formats
- FASTA
- Simple, little annotation information
- Easy to use
- Common denominator format
- ASN1
- Computer friendly, human unfriendly
- XML, INSDSeqXML, TinySeqXML
- Graph (graphical map of seq features)
- and more
20
DNA Sequence Files Common formats
- Genbank (used by VectorNTI)
- FASTA
- GCG
- Accelrys GCG package
- formerly known as the GCG Wisconsin Package
- (GCG Genetics Computer Group)
- Many others!
21
FASTA
One annotation line only!
gtgi1345098gbU30791.1PCU30791
TGAATTCTAAATTTTATATTTCTAATTGCATTTTATATTTTTGATAATAC
TAGATTTATTCCTGGAAACT TAAATTAGTTATTTTAAGTTATGGGATGT
TGTTTTTCTGCTACATATAACCAAGATACACTTCGTTCCAA
22
Submitting sequences to Genbank
- Sequin
- Stand-alone sequence submission tool.
- BankIt
- Web based sequence submission.
23
Genbank is an ARCHIVE
- The literature and secondary databases are the
knowledge sources. - There are many additional NCBI annotation
databases
24
NCBI annotation databases!
- Genbank -gt RefSeq (Single sequence for each gene)
- Entrez Gene (Gene-based links to annotation
sources). - HomoloGene (Homologs)
- OMIM
- Conserved domains, 3D domains
- GEO (Gene expression datasets)
- DNA, protein, 3D structures
- Interaction data
- Links to other databases!
- NCBI Genomes
- NCBI Map viewer
25
Finding and editing DNA files
- Find DNA Entrez
- Downloading files
- Format Conversion
- Sequence viewing/editing
26
Entrez
- (Relational database manager)
- Database searching/browsing
- Example Pneumocystis G-proteins
- PCR a cDNA to express in E. coli
- Read about it and related genes
- Check similarity to related G-proteins
- View the 3D structure??
- http//www.ncbi.nlm.nih.gov/Entrez/
27
Entrez Neighbors-Literature
DNA Protein Structure Genome Popset
Article
Keyword, authors
citation
Article
28
Entrez Neighbors-Protein
3D Structure
citation
citation
Protein
Literature
encoding
BLASTP
Protein
DNA
29
Mapping the menagerie of biological databases
30
Nucleic Acid Manipulations
- VectorNTI
- On the web
- Baylor Human Genome Center (BCM)
http//searchlauncher.bcm.tmc.edu/seq-util/seq-uti
l.html - European Bioinformatics Institute (EBI)
http//www.ebi.ac.uk/Tools/misc.html
31
File Conversion
- Readseq
- Download program
- http//iubio.bio.indiana.edu/soft/molbio/readseq
- Use online
- http//www.ebi.ac.uk/cgi-bin/readseq.cgi
- http//searchlauncher.bcm.tmc.edu/seq-util/readseq
.html
- VectorNTI
- Other utilities
- Readseq ----gt
Beware Information Loss
32
Reverse Complementing
5-GAATCA-3
5-TGATTC-3 NOT 5-ACTAAAG-3
33
Sequence Statistics
- Nucleotide frequencies (di, tri)
- UV Absorbance
- MW
- Tm
34
Restriction Map
- Linear vs Circular
- Enzyme sets
- Which enzymes, where they cut.
- Gel simulation
- Gel-to-map MUCH harder!!
- Useful for
- Cloning
- Southern blots
- Specialized mol bio techniques
35
Translation/ORFs
- Translation table
- Standard vs non-standard
- Frame (1,2,3,4,5,6)
- Segmental translation (exon-intron)
- Primary translation vs mature polypeptide
36
Sequence File Editing
- VectorNTI
- -Windows editor
- (eg Word-save as TEXT)
- Text editor
- Notepad, Simpletext
- Wordprocessor
- vi
MWGTCC IIIIII MWGTCC IIIIII
Nonproportional fonts (courier, monospaced)
37
Primer design program Primer3
http//frodo.wi.mit.edu/cgi-bin/primer3/primer3_ww
w.cgi
38
Primary vs. Derivative Databases
- Primary Databases
- Original submissions by experimentalists
- Content controlled by the submitter
- Examples GenBank, SNP, GEO
- Derivative Databases
- Built from primary data
- Content controlled by third party (NCBI)
- Examples Refseq, TPA, RefSNP, UniGene, NCBI
Protein, Structure, Conserved Domain
39
Other NCBI Databases
- Structure imported structures (PDB)
- Cn3D viewer, NCBI curation
- CDD conserved domain database
- Protein families (COGs and KOGs)
- Single domains (PFAM, SMART, CD)
- dbSNP nucleotide polymorphism
- Gene gene records
- Unifies LocusLink and Microbial Genomes
40
Homologene Cluster
41
Entrez Protein Derivative Database
42
Redundant Proteins
43
RefSeq NCBIs Derivative Sequence Database
- Curated transcripts and proteins
- reviewed
- human, mouse, rat, fruit fly, zebrafish,
arabidopsis - microbial genomes (proteins), and more
- Model transcripts and proteins
- Assembled Genomic Regions (contigs)
- human
- mouse
- rat
- Chromosome records
- Human genome
- microbial
- organelle
- chicken
- honeybee
- sea urchin
- zebrafish
- cow
- dog
- black poplar
srcdb_refseqProperties
ftp//ftp.ncbi.nih.gov/refseq/release/
44
RefSeq Accession Numbers
mRNAs and Proteins NM_123456 Curated
mRNA NP_123456 Curated Protein NR_123456 Curated
non-coding RNA XM_123456 Predicted
mRNA XP_123456 Predicted Protein
XR_123456 Predicted non-coding RNA Gene
Records NG_123456 Reference Genomic
Sequence Chromosome NC_123455 Microbial
replicons, organelle genomes, human
chromosomes Assemblies NT_123456 Contig
NW_123456 WGS Supercontig