Protein translocation into the endoplasmic reticulum ER - PowerPoint PPT Presentation
1 / 28
Title:
Protein translocation into the endoplasmic reticulum ER
Description:
All integral membrane and lumenal proteins of the ER, Golgi complex, plasma ... Identified Sec61p and TRAM, components of the translocon. 35S-methionine ... – PowerPoint PPT presentation
Number of Views:736
Avg rating:3.0/5.0
Title: Protein translocation into the endoplasmic reticulum ER
1
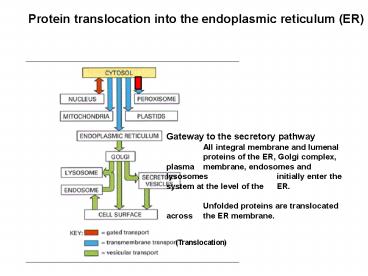
Protein translocation into the endoplasmic
reticulum (ER)
Gateway to the secretory pathway All integral
membrane and lumenal proteins of the ER, Golgi
complex, plasma membrane, endosomes and
lysosomes initially enter the system at the
level of the ER. Unfolded proteins are
translocated across the ER membrane.
(Translocation)
2
Lodish 17-1
3
Protein Sorting Each organelle has a unique
protein composition. (S enzyme activities
function of organelle) All proteins are
synthesized by ribosomes in the cytosol
(except for a few mito. And chloroplast
proteins) This presents a topological and
logistical problem of sorting and delivering
proteins to the appropriate organelle. Signal
Hypothesis Proteins that leave the cytosol have
intrinsic signals that direct them to the
appropriate organelle Gunther Blobel, 1999
Nobel Laureate sorting signals (signals
within the protein that provide its address
label) primary structure (SKL on C-terminus of
peroxisomal proteins) tertiary structure
(soluble lysosomal enzymes) posttranslational
modification (mannose-6-phosphate) trans-acting
components "receptor" that recognizes the
sorting signal sorting machinery that delivers
the protein to appropriate place.
4
Protein translocation into the endoplasmic
reticulum (ER)
Sorting signal Signal sequence or leader
peptide Typically 9 - 15 hydrophobic amino
acids. Near the N-terminus. Upon insertion
into the ER lumen, the signal sequence is
cleaved off by signal peptidase. Defined
experimentally by molecular biology
approaches. Necessary and sufficient
tests. Trans-acting Machinery Defined through
biochemical and genetic approaches Signal
recognition particle (SRP) SRP
receptor Translocon BiP, an ER lumenal Hsp70
chaperone
5
Biochemical approach Required an assay to
reconstitute translocation Cloned cDNA encoding
protein with a signal sequence In vitro
transcribe cDNA to produce a pure mRNA Translate
mRNA using reticulocyte lysate (supplies
ribosomes, charged tRNAs and other cytosolic
factors) ATP 35S-methionine in the presence
or absence of microsomes (purified ER
membranes) Subject samples to SDS-PAGE and
autoradiography to visualize newly synthesized
protein Measure the amount of protein that
translocated into the ER. 1. Signal peptide
cleavage 2. Protease protection 3.
Centrifugation to separate microsomes from
cytosol Purify proteins required for
translocation
6
Most mammalian proteins enter the ER
cotranslationally Some proteins can be
translocated posttranslationally Reflects a
requirement for the protein to be unfolded as it
is translocated
17-15
7
Shields and Blobel, 1978. JBC 2533753
RNA
Reticulocyte lysate
Microsomes
Post-translational addition of protease
Signal cleavage
pregrowth hormone
growth hormone
In this case, the microsomes had to be added at
the same time as the reticulocyte
lysate. Translocation occurred co-translationally.
Some proteins can be translocated after
synthesis.
8
Shields and Blobel, 1978. JBC 2533753
RNA
Reticulocyte lysate
Microsomes
Post-translational addition of protease
Protease
9
Shields and Blobel, 1978. JBC 2533753
RNA
Reticulocyte lysate
Microsomes
Post-translational addition of protease
Protease
10
Purify components from cytosol required for
translocation Signal Recognition Particle (SRP)
Recognizes signal sequences as they emerge from
ribosome Causes a pause in translation Delivers
ribosome with associated mRNA and partially
translated protein to SRP receptor on ER
membrane. GTPase activity of SRP (P54) and the
SRP receptor help drive association/disassociation
of complexes
SRP
11
(No Transcript)
12
Identification of the translocon by chemical
crosslinking
No stop codon so polypeptide remains attached to
ribosome Short polypeptide is synthesized using
a lysyl-tRNA modified with a light-activated
crosslinking reagent Identified Sec61p and TRAM,
components of the translocon
35S-methionine
13
Identification of the translocon by chemical
crosslinking
Gorlich et al 92 Cell 71489
14
Johnson and van Waes, 99 Ann Rev Cell Dev Biol
15799
15
BiP (Hsp70 family protein) helps seal the
translocon from the inside
Johnson and van Waes, 99 Ann Rev Cell Dev Biol
15799
16
http//www.rockefeller.edu/pubinfo/proteintarget.h
tml
Soluble protein
17
Topologies of some integral membrane proteins
The topology of integral membrane proteins is
established During insertion into the ER membrane
and is maintained As the protein is transported
to other membranes
18
Mechanism for generating a Type I integral
membrane protein
19
Mechanism for generating a Type II integral
membrane protein
Uncleaved signal sequence signal-anchor domain
20
(No Transcript)
21
Mechanism for generating a polytopic (or
multispanning) integral membrane protein
Signal-anchor domains are also called
start-transfer sequences
22
Computer programs for prediction of signal
sequences and transmembrane domains
http//us.expasy.org/ Expert Protein Analysis
System
Proteomics and sequence analysis tools
Proteomics PeptIdent, PeptideMass, ...
DNA -gt Protein Translate
Similarity searches BLAST
Pattern and profile searches ScanProsite
Post-translational modification and
topology
prediction
Primary structure analysis ProtParam, pI/MW,
ProtScale
Secondary and tertiary structure prediction
SWISS-MODEL, Swiss-PdbViewer
Alignment T-COFFEE, SIM
Biological text analysis
23
Post-translational modification prediction
SignalP - Prediction of signal peptide cleavage
sites ChloroP - Prediction of
chloroplast transit peptides MITOPROT -
Prediction of mitochondrial targeting sequences
Predotar - Prediction of mitochondrial and
plastid targeting sequences NetOGlyc -
Prediction of type O-glycosylation sites in
mammalian proteins DictyOGlyc -
Prediction of GlcNAc O-glycosylation sites in
Dictyostelium YinOYang - O-beta-GlcNAc
attachment sites in eukaryotic protein sequences
big-PI Predictor - GPI Modification
Site Prediction DGPI - Prediction of
GPI-anchor and cleavage sites (Mirror site)
NetPhos - Prediction of Ser, Thr and Tyr
phosphorylation sites in eukaryotic proteins
NetPicoRNA - Prediction of protease cleavage
sites in picornaviral proteins NMT -
Prediction of N-terminal N-myristoylation
Sulfinator - Prediction of tyrosine sulfation
sites
Topology prediction PSORT - Prediction
of protein sorting signals and localization sites
TargetP - Prediction of subcellular
location DAS - Prediction of
transmembrane regions in prokaryotes using the
Dense Alignment Surface method (Stockholm
University) HMMTOP - Prediction of
transmembrane helices and topology of proteins
(Hungarian Academy of Sciences)
PredictProtein - Prediction of transmembrane
helix location and topology (Columbia University)
SOSUI - Prediction of transmembrane
regions (TUAT Tokyo Univ. of Agriculture
Technology) TMAP - Transmembrane
detection based on multiple sequence alignment
(Karolinska Institut Sweden) TMHMM -
Prediction of transmembrane helices in proteins
(CBS Denmark) TMpred - Prediction of
transmembrane regions and protein orientation
(EMBnet-CH) TopPred 2 - Topology
prediction of membrane proteins (Stockholm
University)
24
Mechanism for generating a glycolipid linked
integral membrane protein
GPI glycosylphosphatidylinositol
25
Protein association with membranes Methods
26
Protease protection assays can be used to assess
protein topology
27
Analysis of Erd1p topology
A. Hydropathy plot using the Kyte and Doolittle
program. Transmembrane domain prediction using
TMHMM program. (J.Mol. Biol. 305 567-580)
B. Epitope tags were added to the N- and C-
termini of Erd1p
C. Cells expressing epitope tagged forms of Erd1
were lysed (L) and the lysates were centrifuged
at 120,000 x g for 1 hr to generate a membrane
pellet (P) and supernatant fraction (S). The
pellets were treated with either Na Carbonate (pH
10.5) or Triton X-100 and centrifuged again to
generate the P and S fractions. The pellets
were also treated with proteinase K in the
presence or absence of detergent. Erd1 in these
sample was examined by Western blot.
D. Predicted topology
28
Output from the TMHMM program for Erd1p
Large red blocks are predicted TMDs, the blue
line indicates predicted cyoplasmic domains and
the pink lines indicate predicted exoplasmic
domains.