Central Dogma of molecular biology - PowerPoint PPT Presentation
1 / 14
Title:
Central Dogma of molecular biology
Description:
Central Dogma of molecular biology The central dogma of molecular biology was first enunciated by Francis Crick in 1958[1] and re-stated in a Nature paper published ... – PowerPoint PPT presentation
Number of Views:656
Avg rating:3.0/5.0
Title: Central Dogma of molecular biology
1
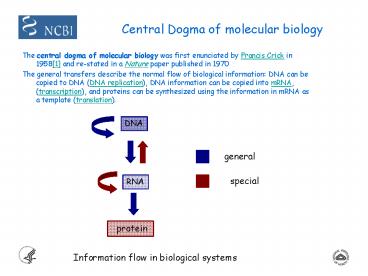
Central Dogma of molecular biology
- The central dogma of molecular biology was first
enunciated by Francis Crick in 19581 and
re-stated in a Nature paper published in 1970 - The general transfers describe the normal flow of
biological information DNA can be copied to DNA
(DNA replication), DNA information can be copied
into mRNA, (transcription), and proteins can be
synthesized using the information in mRNA as a
template (translation).
DNA
general
special
RNA
protein
Information flow in biological systems
2
History of technology
Computer and WWW 1936 Conrad Zuse - first freely
programmable computer automatic-calculator device
would require three basic elements a control, a
memory, and a calculator for the
arithmetic. 1969 Department of Defense developed
ARPANET, the first network that was the beginning
of the Internet. 1990 World Wide Web first
version of hypertext GUI browser
developed by Tim Berners-Lee
Sequencing technology 1973 Sanger capillary
sequencing 1997 Pyrosequencing
3
Capillary electrophoretic sequencing
4
Pyrosequencing or single-molecule method
5
Computing and sequencing
6
Basic Data Model Based on Central Dogma
Resources
Tools
Taxonomy
Organism
GenomeProject
Genomes
Genome
Nucleotide
Chromosome/Contig
Gene
Genes
Transcript
mRNA
Refseq
ProProtein
Protein
Mature peptide
Structure
Structure
CDD
Domains
PubChem
Function
Disease Phenotype
PubMed
PubMed Central
7
Data Resources at NCBI
- Databases Primary and Derivative
- Primary Databases
- Archival submissions of experimental
results - Database staff organize but dont add
additional information - Derivative Databases
- Curated/expert review
- Computationally derived
- Combinations
Genbank
dbEST
dbSTS
dbSNP
Probe
Traces
SRA
Refseq
Genomes
UniGene
Homologene
UniSTS
CDD
Blink
8
Primary sequence Databases
- GenBank/EMBL/DDBJ - author submissions of
nucleotide (genomic and mRNA) sequence with
conceptual translations as appropriate - Sequence Reads Archieve- raw sequence data from
sequencers - dbSTS primers, product sizes, map information
- dbEST sequence data and other information on
"single-pass" cDNA sequences, or Expressed
Sequence Tags - GEO DataSets a high-throughput gene expression /
molecular abundance data repository - Probe a public registry of nucleic acid reagents
designed for use in a wide variety of biomedical
research applications, together with information
on reagent distributors, probe effectiveness, and
computed sequence similarities.
9
Derivative sequence databases
- UniGene-EST sequences clustered by sequence
according to the source gene - UniSTS-PCR primer sequences with predicted
product size, connected to other sequence data by
e-PCR - dbSNP sequence surrounding/including sites of
variation/polymorphism - Assembly Archieve links raw sequence data with
assembly in sequence repository - GEO profiles curated gene expression and
pre-computed profiles - Gene gene-centered information derived form
Refseq - Homologene eukaryotic homology groups
- Protein Clusters protein clusters of orthologous
groups - Related sequences pre-computed BLAST neighbors
- TPA division in GenBank third party annotation
- Protein structure, conserved domains, small
molecular compounds (PubChem) - RefSeq-goal curated representative sequence for
major molecules of the central dogma
10
Primary Data
GenBank
SNP
Data Submission
STS
access
Trace
gt 100,000,000,000 bp
Archival
Gene
Genomes
11
Trace Archive
12
(No Transcript)
13
Assembly Archive
14
(No Transcript)