DNA repair PowerPoint PPT Presentation
1 / 29
Title: DNA repair
1
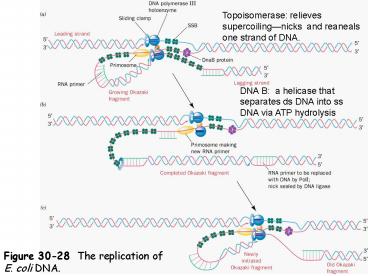
Topoisomerase relieves supercoilingnicks and
reaneals one strand of DNA.
DNA B a helicase that separates ds DNA into ss
DNA via ATP hydrolysis
Figure 30-28 The replication of E. coli DNA.
2
Functional domains in the Klenow Fragment (left)
and DNA Polymerase I (PDB). Produced from
subtilisin or trypsin cleavage Retains
polymerase and 3-5 exo activity
3
Figure 30-8b X-Ray structure of E. coli DNA
polymerase I Klenow fragment (KF) in complex with
a dsDNA (a tube-and-arrow representation of the
complex in the same orientation as Part a).
Page 1141
4
The structure of the Klenow fragment of DNAP I
from E. coli
Fingers
Palm
5
Mismatch repair during DNA replicaiton
6
Replacing RNA primers
7
Nick Translation
- Requires 5-3 activity of DNA pol I
- Steps
- At a nick (free 3 OH) in the DNA the DNA pol I
binds and digests nucleotides in a 5-3
direction - The DNA polymerase activity synthesizes a new DNA
strand - A nick remains as the DNA pol I dissociates from
the ds DNA. - The nick is closed via DNA ligase
Source Lehninger pg. 940
8
Figure 30-20 The reactions catalyzed by E. coli
DNA ligase.
Page 1150
9
Figure 30-21 X-Ray structure of DNA ligase from
Thermus filiformis.
Page 1151
10
Quick Comparison of DNA polymerases I and III Quick Comparison of DNA polymerases I and III Quick Comparison of DNA polymerases I and III
DNA polymerase III DNA polymerase I
Structure asymmetric dimer i. e., two cores with other accessory subunits. It can thus move with the fork and make both leading and lagging strands. monomeric protein, 3 active sites. 5'-to-3' exonuclease and polymerase on the same molecule for removing RNA primers is effective and efficient.
Activities Polymerization and 3'-to-5' exonuclease, but on different subunits. This is the replicative polymerase in the cell. Can only isolate conditional-lethal dnaE mutants. Synthesizes both leading and lagging strands. No 5' to 3' exonuclease activity. Polymerization, 3'-to-5' exonuclease, and 5'-to-3' exonuclease (mutants lacking this essential activity are not viable). Primary function is to remove RNA primers on the lagging strand, and fill-in the resulting gaps.
Vmax (nuc./sec) 250-1,000 nucleotides/second. Only this polymerase is fast enough to be the main replicative enzyme. 20 nucleotides/second. Capable of "filling in" DNA to replace the short (about 10 nucleotides) RNA primers on Okazaki fragments.
Processivity Highly processive. The ß subunit is a sliding clamp. The holoenzyme remains associated with the fork until replication terminates. Pol I does NOT remain associated with the lagging strand, but disassociates after each RNA primer is removed.
Molecules/cell 10-20 molecules/cell. In rapidly growing cells, there are 6 forks. If one processive holoenzyme (two cores) is at each fork, then only 12 core polymerases are needed for replication. About 400 molecules/cell. Higher concentration means that it can reassociate with the lagging strand easily.
11
(No Transcript)
12
DNA Pol III holoenzyme.
13
Figure 30-13b The ? subunit of E. coli Pol III
holoenzyme. Space-filling model of sliding clamp
in hypothetical complex with B-DNA.
Page 1146
14
Sliding clamp
http//www.callutheran.edu/Academic_Programs/Depar
tments/BioDev/omm/poliiib_2/poliiib.htm
15
Heres a computer modelof DNA replicationhttp/
/www.youtube.com/watch?v4jtmOZaIvS0
This is a pretty good outline http//www.youtube.
com/watch?vteV62zrm2P0NR1
Another one with review questions (perhaps
oversimplified) http//www.wiley.com/college/pratt
/0471393878/student/animations/dna_replication/ind
ex.html
16
FIDELITY OF REPLICATION
- Expect 1/103-4, get 1/108-10.
- Factors
- 3?5 exonuclease activity in DNA pols
- Use of tagged primers to initiate synthesis
- Battery of repair enzymes
- Cells maintain balanced levels of dNTPs
17
This article is a simple overview of repair
processes
- http//www.nature.com/nature/journal/v421/n6921/fu
ll/nature01408.html
18
DNA repair
- Ilkka Koskela
- Katri Vilkman
19
Foreword
- DNA
- variation is an essential factor to evolution
(1000-106 lesions per day) - stability is important for the individual (less
than 1/1000 mutations are permanent) - A relatively large amount of genes are devoted to
coding DNA repair functions.
20
- Sources of damage
- heat
- metabolic accidents (free radicals)
- radiation (UV, X-Ray)
- exposure to substances (especially aromatic
compounds)
- Types of damage
- deamination of nucleotides
- depurination of nucleotides
- oxidation of bases
- breaks in DNA strands
21
Diseases
- colon cancer
- cellular ultraviolet sensitivity
- Werner syndrome (premature aging, retarded
growth) - Bloom syndrome (sunlight hypersensitivity)
22
Damage of the double helix
- Single strand damage
- information is still backed up in the other
strand - Double strand damage
- no backup
- can cause the chromosome to break up
23
Single strand repair
- Base excision repair
- A base-specific DNA glycosylase detects an
altered base and removes it - AP endonuclease and phosphodiesterase remove
sugar phosphate - DNA Polymerase fills and DNA ligase seals the
nick
24
Single strand repair
- Nucleotide excision repair
- a large multienzyme compound scans the DNA strand
for anomalities - upon detection a nuclease cuts the strand on both
sides of the damage - DNA helicase removes the oligonucleotide
- the gap is repaired by DNA polymerase and DNA
ligase enzymes
25
Double strand repair
- Nonhomologous end-joining
- only in emergency situations
- two broken ends of DNA are joined together
- a couple of nucleotides are cut from both of the
strands - ligase joins the strands together
26
Double strand repair
- Homologous end-joining
- damaged site is copied from the other chromosome
by special recombination proteins
27
DNA repair enzymes
- a lot of DNA damage -gt elevated levels of repair
enzymes - extreme change in cell's environment (heat, UV,
radiation) activates genes that code DNA repair
enzymes - For an example, heat-shock proteins are produced
in heat-shock response when being subjected to
high temperatures.
28
Cell Cycle and DNA repair
- Cell cycle is delayed if there is a lot of DNA
damage. - Repairing DNA as well as signals sent by damaged
DNA delays progression of cell cycle. - -gtensures that DNA damages are repaired before
the cell divides
29
References
- Pictures
- http//www.2modern.com/index.asp?PageActionVIEWPR
ODProdID985 - http//www.senescence.info/WS.jpg
- http//en.wikipedia.org/wiki/Dna
- http//www.funpecrp.com.br/gmr/year2003/vol1-2/ima
gens/sim0001fig1.jpg - http//www.science.siu.edu/microbiology/micr460/Pa
geMill20Images/image32.gif - http//www.bio.brandeis.edu/haberlab/jehsite/image
s/nhejd.gif - http//www.biochemsoctrans.org/bst/029/0655/bst029
0655f02.gif - http//www.antigenics.com/products/tech/hsp/images
/animation.jpg - http//bioinformatics.psb.ugent.be/images/illust_c
ell_cycle_large.jpg