Sequence Analysis - PowerPoint PPT Presentation
1 / 39
Title:
Sequence Analysis
Description:
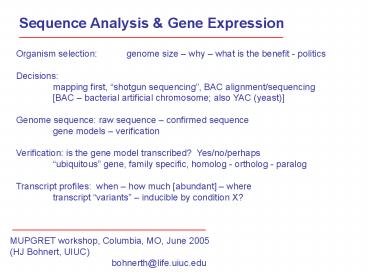
Sequence Analysis & Gene Expression Organism selection: genome size why what is the benefit - politics Decisions: mapping first, shotgun sequencing , BAC ... – PowerPoint PPT presentation
Number of Views:133
Avg rating:3.0/5.0
Title: Sequence Analysis
1
Sequence Analysis Gene Expression
Organism selection genome size why what is
the benefit - politics Decisions mapping
first, shotgun sequencing, BAC
alignment/sequencing BAC bacterial artificial
chromosome also YAC (yeast) Genome sequence
raw sequence confirmed sequence gene models
verification Verification is the gene model
transcribed? Yes/no/perhaps ubiquitous gene,
family specific, homolog - ortholog -
paralog Transcript profiles when how much
abundant where transcript variants
inducible by condition X?
MUPGRET workshop, Columbia, MO, June 2005 (HJ
Bohnert, UIUC) bohnerth_at_life.uiuc.edu
2
Genomics
not just genes
genome transcriptome sequences
protein interaction maps
markers QTLs
ATCCGAAGCG CTTGGAAAA
biochemical genetics
expression profiles
Databases, Integration Intuition
knock-out sRNA RNAi
dynamic metabolite catalogs
protein localization
structure analysis
How (much) will encyclopedic approaches lead
to better understanding?
information mining, hypotheses, experiment -
insight, application, virtual life
3
Arabidopsis model plant small, fast,
prolific, mutants, lines, ecotypes, genome
sequence
Field on a dish!
4
AQP are distributed over all Chromosomes - a few
clusters, many duplications
Arabidopsis thaliana AGI, 2000
5
(No Transcript)
6
The Plant Genome
Plants in silico? Sure! And then Plant
Design from Scratch
7
Controls for Gene Expression many
Switchboards
The Plant Genome
- Chromatin condensation state
- Local chromatin environment
- Transcription initiation
- Transcript elongation
- mRNA splicing
- mRNA export
- mRNA place in the cell
- RNA half-life
- Killer microRNAs
- Ribosome loading
Levels of regulation that affect what we
call gene expression
8
The Plant Transcriptome
5 years ago, we did not know that such a
control system existed!
Killer RNAs (there are micro-genes)
microRNAs
9
The Plant Transcriptome
How to sample the transcriptome?
- Morphological dissection
- (root, leaf, flower - epidermis, guard
cell, etc.) - Cell sorting
- make single cells, send through cell sorter
- (size, color, reporter gene)
- Laser ablation
- micromanipulation of laser to cut
- individual cells
- Biochemical dissection (compartment isolation)
- chloroplasts, mitochondria,
- ribosomes, other membranes
Painting cells with a reporter gene - here
is GFP Green Fluorescence Protein
10
The Plant Transcriptome
Painting tissues then isolating desired cells
Enzymatic staining
The Endodermis of the root tip is highlighted in
transgenic plants using pSCRmGFP5.
Emerging lateral roots
requires plant transformation
11
The Plant Transcriptome
cDNA complementary DNA converts messenger RNA
into double-stranded DNA
- gt cDNA libraries
- neat
- normalized
- subtracted
- gt SAGE libraries
Normalization removes mRNAs for which there
are many copies in a cell thus enriching for
rare mRNAs (not so much sequencing to do)
Subtraction removes cDNAs which you already know
(less sequencing)
12
cDNA Libraries
The Plant Transcriptome
Cloning of root RNAs from segments S1 S4 root
tip (Sharp lab) sequenced 18,000
clones found 8,000 unique and 130 novel
genes How many genes make a root?
13
Serial Analysis Gene Expression
Velculescu et al. 1995
http//www.sagenet.org/
14
coding region (known or expected)
forward p.
reverse p.
Amplicon (sequence or clone sequence)
1 2 3 4 5
6 7 8 9
10 M
results
15
Real-time PCR) (quantitative)
RNA (DNA-free) to cDNA use product in
dilutions for amplification Assumption each
cycle increases amount by factor 2 (or
1.8) Check by using known amount of cloned
control cDNA
cycle number
Serial dilution 1x - 1/5x - 1/25x - 1/125x
16
Melting curves single products
Single genes have been amplified here
Two amplicons are shown Each shows a single
melting curve
17
Melting curves multiple products
More than one gene has been amplified here
Homologous genes identity similarity
divergence
orthologous paralogous relationships
18
The Plant Transcriptome
Quantitative PCR in 384-well plates (96 primer
pairs, 3 repeats each)
Taking SAGE cDNA sequences together - corn
roots express 20-23,000 genes (i.e., mRNA is
made) - The entire corn genome is expected to
include 50,000 genes
19
Substrates for High Throughput Arrays
Single label 33P
Single label biotin streptavidin
Dual label Cy3, Cy5
20
TeleChem ChipMaker2 Pins
21
Creating cDNA Arrays
Q-Pix
cDNA cloned into vector and transformed to create
cDNA library
Clones sequenced and unique set chosen and
reracked
Unique set of clones
384 well microtiter plate
Slides printed on Cartesian Arrayer
PCR on Tecan workstation
Final product
22
Printing Arrays on 50 slides
23
Slide Chemistry
Glass
Coatings
Amine
Poly-L-lysine
Silanated
We use SuperAmine and SuperAldehyde from
TeleChem (arrayit.com)
24
GSI Lumonics
25
GenePix Image Analysis Software
Placenta vs. Brain 3800 Cattle Placenta Array
cy3 cy5
26
The Good
The Ugly
The Bad
27
Post-Print Processing
Hot
Printed slide
Snap dry
W
ater
Rehydrate spots
UV light
Chemically block background. Denature to single
strands.
Hybridize Scan
Fix DNA to coating
28
Ratio of expression of genes from two sources
29
ScanArray 3000 Fluorescent Scanner
30
Overlay Images
Reverse Labeling
Slide 2 Cy5 over-expressed
Slide 1 Cy3 over-expressed
31
Universal vs. Universal (control v. control)
Problem area at low intensity readings
32
Lung vs Control
33
Clustered display of data from time course of
serum stimulation of primary human fibroblasts.
Cholesterol Biosynthesis
Cell Cycle
Immediate Early Response
Signaling and Angiogenesis
Wound Healing and Tissue Remodeling
Eisen et al. Proc. Natl. Acad. Sci. USA 95
(1998) pg 14865
34
Hierarchical Clustering 14 Tissues 7653
Genes
35
Differences in Technology
Affymetrix
- One sample, one chip
- Single Color Scans
- Labeling by incorporating Biotin into cRNA
not Cy3 or Cy5 dyes - Oligonucleotides instead of full-length cDNAs
- Higher Density Arrays
- Feature sizes down to 18 mm instead of 100 mm
- Non-contact Creation of Arrays
GeneChips
36
Affy Technology Overview
- Photolithography and combinatorial chemistry
- Technology from microchip industry GeneChip
- Coat slides
- Mask to apply light to only desired features,
de-protects feature
37
Technology Overview (cont.)
- Apply required nucleotide base to array
- Apply new mask to de-protect different features
- Stack nucleotides on top of one another
- Repeat with bases and masks until 25-mer
oligonucleotides are built directly onto array
38
Technology Final Steps
- Silicon wafers of 90 arrays are cut
- Glass substrate is then added to plastic
cartridge for - Safe handling
- Easy storage
- Easy hybridization
- Easy scanning
Easy, convenient Expensive (very much so) No
confirmation of quality Erroneous data when low
intensity Problems with SNPs
not with 70-mer oligo glass slides
39
Questions? Give me a call or send a message
217-265-5475 bohnerth_at_life.uiuc.edu
http//www.life.uiuc.edu/bohnert/ Rem
ember YOU CAN ALWAYS FIND EVERYTHING ON
GOOGLE! (though not these slides)