Sandeepa Chauhan, Madhumita Roy Chowdhury, Neerja Gupta, Sheffali Gulati*,
1 / 1
Title:
Sandeepa Chauhan, Madhumita Roy Chowdhury, Neerja Gupta, Sheffali Gulati*,
Description:
Sandeepa Chauhan, Madhumita Roy Chowdhury, Neerja Gupta, Sheffali Gulati*, BK Thelma#, Anjali Dabral#, Madhulika Kabra Genetic Unit, Department of Pediatrics ... –
Number of Views:56
Avg rating:3.0/5.0
Title: Sandeepa Chauhan, Madhumita Roy Chowdhury, Neerja Gupta, Sheffali Gulati*,
1
Introduction
New approach to fragile X analysis
Sandeepa Chauhan, Madhumita Roy Chowdhury, Neerja
Gupta, Sheffali Gulati, BK Thelma, Anjali
Dabral, Madhulika Kabra Genetic Unit,
Department of Pediatrics, Department of
Pediatrics, AIIMS, New Delhi Department of
Genetics, University of Delhi South Campus, New
Delhi
Abstract
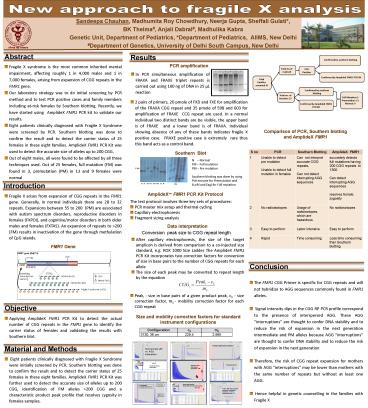
Results
Confirmed by southern blotting
PCR amplification
- Fragile X syndrome is the most common inherited
mental impairment, affecting roughly 1 in 4,000
males and 1 in 7,000 females, arising from
expansion of CGG repeats in the FMR1 gene. - Our laboratory strategy was to do initial
screening by PCR method and to test PCR positive
cases and family members including at-risk
females by Southern blotting. Recently, we have
started using AmplideX FMR1 PCR kit to validate
our results. - Eight patients clinically diagnosed with Fragile
X Syndrome were screened by PCR. Southern
blotting was done to confirm the result and to
detect the carrier status of 25 females in these
eight families. AmplideX FMR1 PCR Kit was used to
detect the accurate size of alleles up to 200
CGG. - Out of eight males, all were found to be affected
by all three techniques used. Out of 25 females,
full mutation (FM) was found in 3, premutation
(PM) in 13 and 9 females were normal
Total no.of males8
PCR Positive
- In PCR simultaneous amplification of FRAXA and
FRAXE triplet repeats is carried out using 100 ng
of DNA in 25 µL reaction
Confirmed by AmplideX FMR1 PCR Kit
Total families screened 8
Confirmed by southern blotting
Total no. of females 25
Full Mutation3 Premutation 13 Normal 9
- 2 pairs of primers, 20 pmole of FXD and FXE for
amplification of the FRAXA CGG repeat and 35
pmole of 598 and 603 for amplification of FRAXE
CCG repeat are used. In a normal individual two
distinct bands are be visible, the upper band is
of FRAXE and a lower band is of FRAXA.
Individual showing absence of any of these bands
indicates fragile X positive case. FRAXE
positive case is extremely rare thus this band
acts as a control band.
Confirmed by AmplideX FMR1 PCR Kit
Comparison of PCR, Southern blotting and
AmplideX FMR1
S no PCR Southern Blotting AmplideX FMR1
1 Unable to detect pre mutation Unable to detect full mutation in females Can not interpret accurate CGG repeats, Can not detect interrupting AGG sequences accurately detects full mutations having 200 CGG repeats to 1300 Can detect interrupting AGG sequences resolves female zygosity
2 No radioisotopes Usage of radioisotopes, which are hazardous No radioisotopes
3 Easy to perform Labor intensive Easy to perform
4 Rapid Time consuming Less time consuming than Southern blotting
Southern Blot
N Normal FM Full mutation PM Pre
mutation Southern blotting was done by using
PstI enzyme for Premutation and EcoRI and EagI
for Full mutation
5.2Kb
PM
AmplideX FMR1 PCR Kit Protocol
- Fragile X arises from expansion of CGG repeats in
the FMR1 gene. Generally, in normal individuals
there are 28 to 32 repeats. Expansions between 55
to 200 (PM) are associated with autism spectrum
disorders, reproductive disorders in females
(FXPOI), and cognitive/motor disorders in both
older males and females (FXTAS). An expansion of
repeats to gt200 (FM) results in inactivation of
the gene through methylation of CpG islands.
- The test protocol involves three key sets of
procedures - PCR master mix setup and thermal cycling
- Capillary electrophoresis
- Fragment sizing analysis
Data interpretation
Conversion peak size to CGG repeat length
- After capillary electrophoresis, the size of the
target amplicon is derived from comparison to a
co-injected size standard, e.g. ROX 1000 Size
Ladder The AmplideX FMR1 PCR Kit incorporates two
correction factors for conversion of size in base
pairs to the number of CGG repeats for each
allele - The size of each peak may be converted to repeat
length by the equation
FMR1 Gene
Conclusion
- The FMR1 CGG Primer is specific for CGG repeats
and will not hybridize to AGG sequences commonly
found in FMR1 alleles. - Signal intensity dips in the CGG RP PCR profile
correspond to the presence of interspersed AGG.
These AGG interruptions are thought to confer
DNA stability and to reduce the risk of expansion
in the next generation intermediate and PM
alleles because AGG interruptions are thought
to confer DNA stability and to reduce the risk of
expansion in the next generation - Therefore, the risk of CGG repeat expansion for
mothers with AGG interruptions may be lower
than mothers with the same number of repeats but
without at least one AGG. - Hence helpful in genetic counselling in the
families with Fragile X
- Peaki - size in base pairs of a given product
peak, c0 - size correction factor, m0 - mobility
correction factor for each CGG repeat
Objective
Size and mobility correction factors for
standard instrument configurations
- Applying AmplideX FMR1 PCR Kit to detect the
actual number of CGG repeats in the FMR1 gene to
identify the carrier status of females and
validating the results with Southern blot.
Configuration c0 m0
3130, 36 cm 229.4 2.965
Material and Methods
Female with two normal X
Normal male with no AGG interruption
- Eight patients clinically diagnosed with Fragile
X Syndrome were initially screened by PCR.
Southern blotting was done to confirm the result
and to detect the carrier status of 25 females in
these eight families. AmplideX FMR1 PCR Kit was
further used to detect the accurate size of
alleles up to 200 CGG, identification of FM
alleles gt200 CGG and a characteristic product
peak profile that resolves zygosity in females
samples.
AGG interruption in both X in 2 locations
29 CGG
Premutation78 CGG
Male FM mosaic with all peak populations gt200
Female 29/29 CGG homozygous
Female heterozygous Normal/FM with 2 AGG
interruption
AGG interruption
125 CGG
Premutation Female 108-125 CGG
FM gt200
Two X, with one normal X and one pre-mutation
mosaic X. 2 AGG interruptions on 1 X
108 CGG