Sequence database searching - PowerPoint PPT Presentation
Title:
Sequence database searching
Description:
Title: PowerPoint Presentation Author: heringa Last modified by: heringa Created Date: 2/20/2003 6:08:47 PM Document presentation format: On-screen Show – PowerPoint PPT presentation
Number of Views:31
Avg rating:3.0/5.0
Title: Sequence database searching
1
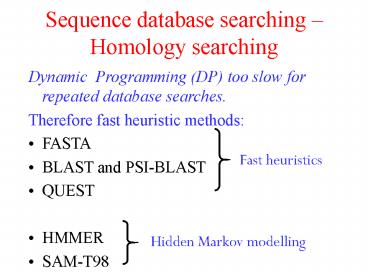
Sequence database searching Homology searching
- Dynamic Programming (DP) too slow for repeated
database searches. - Therefore fast heuristic methods
- FASTA
- BLAST and PSI-BLAST
- QUEST
- HMMER
- SAM-T98
Fast heuristics
Hidden Markov modelling
2
FASTA
- Compares a given query sequence with a library of
sequences and calculates for each pair the
highest scoring local alignment - Speed is obtained by delaying application of the
dynamic programming technique to the moment where
the most similar segments are already identified
by faster and less sensitive techniques - FASTA routine operates in four steps
3
FASTA
- Operates in four steps
- Rapid searches for identical words of a user
specified length occurring in query and database
sequence(s) (Wilbur and Lipman, 1983, 1984). For
each target sequence the 10 regions with the
highest density of ungapped common words are
determined. - These 10 regions are rescored using Dayhoff
PAM-250 residue exchange matrix (Dayhoff et al.,
1983) and the best scoring region of the 10 is
reported under init1 in the FASTA output. - Regions scoring higher than a threshold value and
being sufficiently near each other in the
sequence are joined, now allowing gaps. The
highest score of these new fragments can be found
under initn in the FASTA output. - full dynamic programming alignment (Chao et al.,
1992) over the final region which is widened by
32 residues at either side, of which the score is
written under opt in the FASTA output.
4
FASTA output example
DE METAL RESISTANCE PROTEIN YCF1 (YEAST CADMIUM
FACTOR 1). . . . SCORES Init1 161 Initn 161
Opt 162 z-score 229.5 E() 3.4e-06
Smith-Waterman score 162 35.1 identity in 57
aa overlap
10 20 30 test.seq
MQRSPLEKASVVSKLFFSW
TRPILRKGYRQRLE
YCFI_YEAST CASILLLEALPKKPLMPHQHIHQTLTRRKPNPY
DSANIFSRITFSWMSGLMKTGYEKYLV 180
190 200 210 220 230
40 50 60
test.seq LSDIYQIPSVDSADNLSEKLEREWDRE
YCFI_YEAST EADLYKLPRNFSSEELSQKLEKNWENELKQKSN
PSLSWAICRTFGSKMLLAAFFKAIHDV 240
250 260 270 280 290
5
FASTA
- (1) Rapid identical word searches
- Searching for k-tuples of a certain size within a
specified bandwidth along search matrix
diagonals. - For not-too-distant sequences (gt 35 residue
identity), little sensitivity is lost while speed
is greatly increased. - Technique employed is known as hash coding or
hashing a lookup table is constructed for all
words in the query sequence, which is then used
to compare all encountered words in each database
sequence.
6
FASTA
- The k-tuple length is user-defined and is usually
1 or 2 for protein sequences (i.e. either the
positions of each of the individual 20 amino
acids or the positions of each of the 400
possible dipeptides are located). - For nucleic acid sequences, the k-tuple is 5-20,
and should be longer because short k-tuples are
much more common due to the 4 letter alphabet of
nucleic acids. The larger the k-tuple chosen, the
more rapid but less thorough, a database search.
7
BLAST
- blastp compares an amino acid query sequence
against a protein sequence database - blastn compares a nucleotide query sequence
against a nucleotide sequence database - blastx compares the six-frame conceptual protein
translation products of a nucleotide query
sequence against a protein sequence database - tblastn compares a protein query sequence against
a nucleotide sequence database translated in six
reading frames - tblastx compares the six-frame translations of a
nucleotide query sequence against the six-frame
translations of a nucleotide sequence database.
8
BLAST
- Generates all tripeptides from a query sequence
and for each of those the derivation of a table
of similar tripeptides number is only fraction
of total number possible. - Quickly scans a database of protein sequences for
ungapped regions showing high similarity, which
are called high-scoring segment pairs (HSP),
using the tables of similar peptides. The initial
search is done for a word of length W that scores
at least the threshold value T when compared to
the query using a substitution matrix. - Word hits are then extended in either direction
in an attempt to generate an alignment with a
score exceeding the threshold of S, and as far as
the cumulative alignment score can be increased.
9
BLAST
- Extension of the word hits in each direction are
halted - when the cumulative alignment score falls off by
the quantity X from its maximum achieved value - the cumulative score goes to zero or below due to
the accumulation of one or more negative-scoring
residue alignments - upon reaching the end of either sequence
- The T parameter is the most important for the
speed and sensitivity of the search resulting in
the high-scoring segment pairs - A Maximal-scoring Segment Pair (MSP) is defined
as the highest scoring of all possible segment
pairs produced from two sequences.
10
PSI-BLAST
- Query sequences are first scanned for the
presence of so-called low-complexity regions
(Wooton and Federhen, 1996), i.e. regions with a
biased composition likely to lead to spurious
hits are excluded from alignment. - The program then initially operates on a single
query sequence by performing a gapped BLAST
search - Then, the program takes significant local
alignments found, constructs a multiple alignment
and abstracts a position specific scoring matrix
(PSSM) from this alignment. - Rescan the database in a subsequent round to find
more homologous sequences Iteration continues
until user decides to stop or search has
converged
11
PSI-BLAST iteration
Query sequence
Q
xxxxxxxxxxxxxxxxx
Gapped BLAST search
Query sequence
Q
xxxxxxxxxxxxxxxxx
Database hits
A C D . . Y
PSSM
Pi Px
Gapped BLAST search
A C D . . Y
PSSM
Pi Px
Database hits
12
PSI-BLAST output example
13
Multiple alignment profiles Gribskov et al.
1987A way to represent multiple alignment
consensus
i
A C D ? ? ? W Y
0.3 0.1 0 ? ? ? 0.3 0.3
Gap penalties
0.5
1.0
Position dependent gap penalties
14
Normalised sequence similarity
The p-value is defined as the probability of
seeing at least one unrelated score S greater
than or equal to a given score x in a database
search over n sequences. This probability
follows the Poisson distribution (Waterman and
Vingron, 1994)
P(x, n) 1 e-n?P(S? x), where n is the
number of sequences in the database Depending on
x and n (fixed)
15
Normalised sequence similarityStatistical
significance
The E-value is defined as the expected number of
non-homologous sequences with score greater than
or equal to a score x in a database of n
sequences E(x, n)
n?P(S ? x) if E-value 0.01, then the expected
number of random hits with score S ? x is 0.01,
which means that this E-value is expected by
chance only once in 100 independent searches over
the database. if the E-value of a hit is 5, then
five fortuitous hits with S ? x are expected
within a single database search, which renders
the hit not significant.
16
Normalised sequence similarityStatistical
significance
- Database searching is commonly performed using an
E-value in between 0.1 and 0.001. - Low E-values decrease the number of false
positives in a database search, but increase the
number of false negatives, thereby lowering the
sensitivity of the search.
17
HMM-based homology searching
- Most widely used HMM-based profile searching
tools currently are SAM-T98 (Karplus et al.,
1998) and HMMER2 (Eddy, 1998) - formal probabilistic basis and consistent theory
behind gap and insertion scores - HMMs good for profile searches, bad for alignment
- HMMs are slow
18
The HMM algorithms
- Questions
- What is the most likely die (predicted) sequence?
Viterbi - What is the probability of the observed sequence?
Forward - What is the probability that the 3rd state is B,
given the observed sequence? Backward
19
HMM-based homology searching
Transition probabilities and Emission
probabilities Gapped HMMs also have insertion
and deletion states
20
Profile HMM mmatch state, I-insert state,
ddelete state go from left to right. I and m
states output amino acids d states are silent.
21
Homology-derived Secondary Structure of Proteins
(HSSP) Sander Schneider, 1991