Pr PowerPoint PPT Presentation
Title: Pr
1
Native and Denaturing Mass Spectrometry (MS) in
the context of Structural Genomics
H. Nierengarten1,2, M. Ruff1, A. Poterszman1, I.
Billas1, N. Rochel1, N. Potier3, M. Argentini2,
S. Sanglier3, J.C. Thierry1, D. Moras1, A. Van
Dorsselaer3
- 1 Structural Biology and Genomics Dept., IGBMC,
Illkirch, France 2. MALDI-TOF Facility IGBMC,
Illkirch Strasbourg, France - 3. Dept. of Bio-Organic Mass Spectrometry
(LSMBO), Strasbourg, FRANCE
The two main mass spectrometers dedicated to the
study of proteins of the IGBMC proteomic open
platform
1)The ESI-TOF mass spectrometer (Micromass,
Waters) for native (physiologic conditions) and
denaturing (acidic conditions) MS studies.
2) The MALDI-TOF mass spectrometer (Reflex IV,
Bruker) to identify proteins by peptide mass
finger printing.
Abstract The use of molecular MS, for the
characterization of biomacromolecules is now well
established MALDI-MS and ESI-MS under denaturing
conditions provide information on the molecular
masses, on the purity and on primery sequences of
proteins. More recently, thanks to the gentle
nature of the ESI process, specific
non-denaturing ESI (native or supramolecular MS)
protocols have been developed to detect and
investigate non-covalent interactions (i.e.
secondary, tertiary, and quaternary structures of
protein complexes). In the field of Structural
Genomics at IGBMC, MS is now routinely used to
assist structure determinations through each
stage of the process of protein crystallography
or NMR. Here, we summarize the multiple
possibilities of Denaturing and Native MS in a
Structural Genomics context.
2) Native Mass Spectrometry (supramolecular MS)
Sample preparation The sample preparation is one
of the key step for a good MS result. Prior to
any mass analysis, the sample has to be desalted
(generally on centricon micro-concentrtors)
against ammonium acetate. Ammonium acetate
enables native structures of proteins to be
preserved and is compatible with ESI-MS analyses.
Typically between 3 and 6 dilution/concentration
steps are performed. After desalting, the
protein is diluted to 5 pmol/mL in a 11
water-acetonitrile mixture acidified with 1
formic acid for a denaturing MS experiment . For
native experiments, samples are diluted to 20
pmol/mL in ammonium acetate (20-50 mM, pH 6.8).
a) Possibilities of native MS in a structural
genomics context ?
- Characterization and study of specific
protein/protein, protein/ ligand,
protein/peptide, protein/DNA, protein/metal
non-covalent complexes..
-Determination of the stoichiometry of
interaction.
1) Denaturing Mass Spectrometry (molecular MS)
-Screening libraries of ligands.
- Detection of fortuitous ligands captured during
expression and/or purification (see exemple 2).
2
b) Ex 2 detection of an unexpected ligand in a
NR using denaturing and native ESI-MS
a) Possibilities of denaturing MS in a structural
genomics context ?
- Characterize (in terms of sequence integrity) and
evaluate the purity (or homogeneity) of protein
(or oligonucleotide) preparations to verify that
the desired protein construct has been correctly
expressed. If the molecular weight is not in
agreement with the theoretical one, the protein
is analyzed by MALDI-MS to identify the
unexpected modification and/or deletion using
peptide mass finger printing.
- Direct identification of macromolecular crystal
content not easely detectable by other classical
techniques to verify if the protein has
crystallized with all its expected partners 1.
-Verify the successful incorporation of
seleno-methionine (see exemple 1) or stable
isotopes (N15C13)
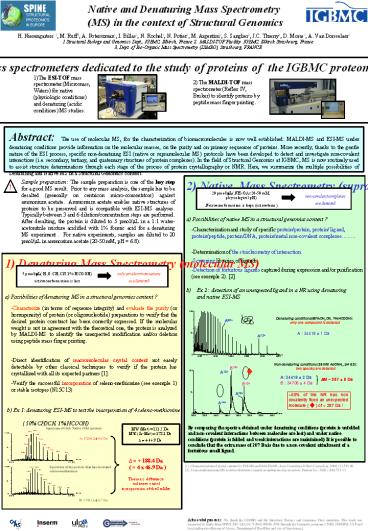
b) Ex 1 denaturing ESI-MS to test the
incorporation of 4 seleno-methionine ( 50 CH3CN,
1 HCOOH)
By comparing the spectra obtained under
denaturing conditions (protein is unfolded and
non-covalent interactions between molecules are
lost) and under native conditions (protein is
folded and weak interactions are maintained) It
is possible to conclude that the extra mass of
287 Da is due to a non-covalent attachment of a
fortuitous small ligand.
Spectrum of theh Native (Met) protein
MW (Met) 131.2 Da MW (Se-Met) 178.1 Da
A 17263.2 0.6 Da
D 46.9 Da
- 188.4 Da
- ( 4 x 46.9 Da )
1. Characterization of crystal content by
ESI-MS and MALDI-MS, Acta Crystallogr D Biol
Crystallogr, 2000 121583-90. 2. Using
nondenaturing MS to detect fortuitous ligands in
orphan nuclear receptors, Protein Sci., 2003
12(4)725-33.
Spectrum of the protein that has incorated
seleno-methionines
The mass difference indicates a
total incorporation of the Se-Met
B 17451.6 0.7 Da
Acknowledgements We thank the LSMBO and the
Structural Biology and Genomics Dept. members.
This work was supported by funds from SPINE EEC
QLG2-CT-2002-00988, FNS through the Genopole
program, CNRS, INSERM, ULP and local authorities
(Region of Alsace, Department of Bas-Rhin and
city of Strasbourg).