Translation PowerPoint PPT Presentation
1 / 34
Title: Translation
1
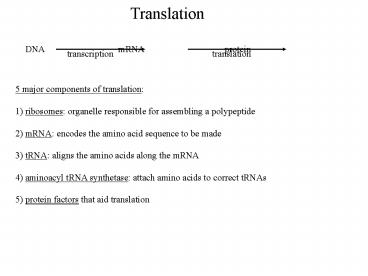
Translation
DNA mRNA
protein
transcription
translation
5 major components of translation 1) ribosomes
organelle responsible for assembling a
polypeptide 2) mRNA encodes the amino acid
sequence to be made 3) tRNA aligns the amino
acids along the mRNA 4) aminoacyl tRNA
synthetase attach amino acids to correct
tRNAs 5) protein factors that aid translation
2
Translation
ribosomes organelle made of rRNA and proteins
composed of 2 subunits, one large, one small
assembled in the nucleolus of the nucleus
found in the cytoplasm, on ER, chloroplasts, and
mitochondria
3
Translation
4 key functional sites in the ribosome located
between the 2 subunits 1) mRNA binding site 2)
A site-- binds each newly arriving tRNA 3) P
site-- site for tRNA with the growing
polypeptide chain 4) E (exit) site-- site where
tRNA leave the ribosome after they transfer
their amino acid
4
tRNA in translation
there is no obvious reason for a codon to be read
as an amino acid requires some kind of adaptor--
must recognized mRNA sequence AND have a link
to one particular amino acid tRNA was shown as a
small RNA found covalently bound to amino
acids 3' OH of adenine in all tRNAs binds the
amino acid 'anticodon' loop is always on the
same position of the tRNA anticodon loop is
complimentary to the mRNA codon
3' OH
amino acid attachment site
5'
stem
anticodon loop
anticodon
5
tRNA in translation
tRNAs are named by superscripts of the amino
acids they carry tRNAala tRNA for alanine,
tRNAser tRNA for serine aminoacyl tRNA tRNA
with an amino acid attached tRNA with an amino
acid is referred to as charged amino acid is
referred to as activated (as in activated
monomer) anti-codon is the 3 base group of the
tRNA which recognizes the codon complimentary
to the codon codons are always listed 5' to 3'
(ie. direction of mRNA synthesis) anti-codons
are listed 3' to 5' (to fit codons as the
complimentary strand) ie. if the codon is
5'-AUG-3' , the anti-codon is 3'-UAC-5'
6
tRNA in translation
of 64 possible codons, 61 code for amino acids
there are fewer than 61 tRNAs-- therefore some
must be reused wobble hypothesis theory that
when the tRNAs line up along the mRNA in a
ribosome there is a fair amount of flexibility in
the third base pair fits the fact that several
codons code for the same amino acid also depends
upon base modification of tRNAs inosine an
unusual nucleic acid that can base pair with most
other bases
7
Aminoacyl-tRNA Synthetases
aminoacyl-tRNA synthetase class of enzymes which
join particular amino acids to the correct tRNA
each amino acid is added by a specific tRNA
synthetase joining reaction uses ATP going to
AMP during aminoacyl-tRNA synthesis bond
between tRNA and amino acid is a 'high energy'
bond-- drives peptide bond formation
tRNA- synthetase recognition areas
amino acid tRNA ATP
aa-tRNA AMP PP
8
Translation
aminoacyl-tRNA synthetases recognize both the
anti-codon region as well as the 3' end of the
tRNA molecule aminoacyl-tRNA is the molecule
that recognizes a codon in mRNA mRNAs also have
to be recognized by the ribosome mRNA leader
sequence of mRNA which is not translated into
protein but rather serves to position the
ribosome to start translation start codon
typically AUG (methionine) for most mRNAs mRNA
trailer sequence of mRNA that follows the stop
codon regulates mRNA stability protein
factors that recognize mRNA leaders, trailers,
and other sequences regulate initiation,
elongation, and termination of translation
9
Mechanism of Translation
start codon is the most 5' codon of the mRNA that
is translated start codon makes up the
N-terminus (free amine group) of the
protein protein synthesis moves from the
N-terminus to the C-terminus 5' to 3' along
the mRNA protein synthesis is divided into 3
distinct stages 1) initiation-- making the
first peptide bonds between amino acids 2)
elongation-- sequentially adding additional amino
acids 3) termination-- stopping protein
synthesis and freeing the chains
10
Initiation of Translation
i) 3 initiation factors (IF1,2, and 3) bind to
the small ribosome subunit IF2 is bound by GTP
(similar to ATP) ii) small ribosome with
initiation factors binds mRNA at a particular
RNA sequence, the Shine-Dalgarno sequence,
displacing IF1 and IF3 (IF2 remains bound for
now), and the initiator tRNA binds in the P
site of the ribosome initiator tRNA codes for
N-formyl methionine, a modified amino acid
forces the charged tRNA into the P site of the
ribosome, not the A site iii) large ribosome
subunit binds to the small subunit-mRNA complex
IF2 is released and GTP is hydrolyzed to GDP at
the end of initiation, ribosome subunits are
assembled with a mRNA positioned between them
and a charged tRNA at the initiator codon
there is a free A site and a free E (exit) site
on the ribosome
11
(No Transcript)
12
Initiation of Translation
eukaryotes have eIFs with eIF2 recognizing the
initiator tRNA before associating with the
small ribosome subunit other proteins recognize
the polyA tail and the 5' mRNA cap
13
Protein Chain Elongation
elongation is a repetitive 3 step cycle 1)
binding of a charged tRNA complimentary to the
next mRNA codon at the A site of the
ribosome 2) formation of a peptide bond between
the carboxyl group of the amino acid at the P
site (separating the amino acid from the tRNA) to
the free amino group of the charged tRNA in
the A site 3) translocation of the ribosome
along the mRNA ratchet movement of mRNA 3
codons so that the free tRNA in the P site
moves to the E site (where it leaves), the
charged tRNA (with attached polypeptide) moves
to the P site, and the next codon (3' of the
last) moves to the A site for the addition of the
next charged tRNA
14
Protein Chain Elongation
Step 1 binding of aminoacyl tRNA to the A site
on the ribosome requires the correct charged
tRNA complimentary to the codon requires 2
protein elongation factors EF-Tu and EF-Ts and
2 GTP converting to GDP (energetically
expensive!) EF-Tu generates the binding of the
tRNA at the A site hydrolyzing GTP EF-Ts
regenerates EF-Tu(GDP) to EF-Tu(GTP) EF-Tu
interacts with any charged tRNA-- not
specific GTP is hydrolyzed only when the tRNA
codon is complimentary error rate for amino acid
incorporation approximately 1 in 10,000
15
Protein Chain Elongation
Step 2 Peptide bond formation carboxyl group
of the elongating chain is attached to the tRNA
at the P site free amino group of the charged
tRNA at the A site peptidyl transferase
enzymatic activity associated with peptide bond
formation-- carried out by a rRNA in the large
ribosomal subunit joins the carboxyl end of
the amino acid at the P site to the amino end
of the amino acid at the A site A site
methionine is always at the N terminus of a newly
made protein energy for peptide bond formation
comes from breaking the bond between the amino
acid and the tRNA in the P site the charged
tRNA was a high energy bond-- ATP AMP
16
Protein Chain Elongation
Step 3 Translocation requires EF-G
elongation factor and GTP (energy driving
translocation) ribosome moves 3 nucleotides (1
codon) along the mRNA transcript free tRNA moves
from the P site to the E (exit) site free tRNA
leaves the ribosome/mRNA complex to get
recharged charged tRNA (with polypeptide) moves
from the A site to the P site A site becomes
free for the next charged tRNA growing
polypeptide chain passes through a 'tunnel' in
the large subunit mRNA goes out from between the
ribosomal subunits 3 steps occur very rapidly--
40 amino acids added per second!
17
Protein Chain Elongation
binding
peptide bond
binding
translocation
18
Translation Termination
eventually a termination codon of the mRNA
reaches the A site no tRNAs recognize stop
codons-- recognized by release factor
proteins release factors mimic tRNA-- charged
tRNA bond is hydrolyzed polypeptide chain
(with free carboxyl group) is released from
ribosome ribosomal subunits come apart, freeing
mRNA and allowing next mRNA and ribosomes to
bind every amino acid requires a lot of energy
to be incorporated ATP AMP to
charge the tRNA 2x GTP GDP to bring
the charged tRNA to the A site GTP
GDP to translocate the ribosome along the
mRNA added to the energy to make mRNA, protein
synthesis takes up a large fraction of the
energy cost of the cell
19
Nonsense Mutations
normally, there is no tRNA that binds to a stop
codon (release factor is a protein) some
bacteria have a mutated tRNA that can bind to one
or more stop codons and add an amino acid
according to the normal elongation
steps suppressor tRNA tRNA that can bind to
stop codons and allow protein synthesis to
continue suppressor tRNAs are usually low
efficiency, allowing most proteins to stop
normally Allows bacteria to tolerate some
mutations that would normally cause proteins
to be truncated (ie. short)
20
Polypeptide Processing
Proteins coming off the ribosome are not
completely finished most proteins lose the
methionine amino acid that initiates
translation proteins become phosphorylated,
methylated, glycosylated, etc posttranslational
processing general term for any chemical
modifications to a polypeptide chain that
occur after translation proteins also need to
get to the location in the cell where they can
function
21
Protein Targeting and Sorting
in eukaryotes, proteins go to specific
organelles-- ie. nucleus, cell membrane,
mitochondria, etc. specific signals must direct
proteins to their appropriate locations can be
divided into 3 separate types of locations 1)
cytoplasm-- general proteins 2) membranes--
nuclear membrane, cell membrane, ER, golgi,
etc 3) membrane bound organelles (mitochondria,
chloroplasts, etc)
22
Protein Targeting and Sorting
all mRNAs initiate translation as free
ribosomes-- ie. small subunits bind the
initiation factors, mRNA, recruits the large
ribosomal subunit as the polypeptide starts
leaving the ribosome, proteins recognize a new
protein chain being synthesized molecular
chaperones proteins which aid (catalyze)
protein folding often bind to proteins as they
are synthesized protein folding into
secondary/tertiary structures occurs WHILE the
polypeptide is being synthesized for membrane
proteins going through the endoplasmic reticulum,
they all start with an ER signal sequence, a
protein sequence that directs import into the
endoplasmic reticulum ER signal sequence gets
recognized, and that ribosome/mRNA becomes
'docked' to the endoplasmic reticulum (ie.
becomes 'rough')
23
Protein Targeting and Sorting
24
Protein Targeting and Sorting
cotranslational import process of coupling
protein synthesis to ER importation-- growing
chain is directly threaded into the
ER N-terminal ER signal sequence is generally
cleaved from the imported protein after it is
imported, generally 15-30 amino acids
long signal sequence has 3 parts N-terminal
charged region, hydrophobic section, and a
polar region adjacent to the cleavage
site signal recognition particle (SRP) protein
complex which recognizes the ER signal
sequence on a free ribosome and attaches to the
ER composed of 6 proteins and 1 RNA
molecule SRP binds to a translocon special
structure in the ER where translocation across
the membrane occurs translocon includes an SRP
receptor, ribosome receptor, a pore through
the membrane, and a signal peptidase to cleave
the ER signal sequence
25
Protein Targeting and Sorting
6 steps of protein translocation into the ER 1)
SRP binds growing polypeptide chain on a free
ribosome 2) SRP binds to the SRP receptor on the
endoplasmic reticulum 3) GTP binds to the
SRP/SRP receptor and causes transfer of the
polypeptide through the translocon 4) GTP is
hydrolyzed to GDP and SRP comes off 5) peptide
loops into the ER while the signal sequence
remains bound 6) signal peptidase cleaves the
signal sequence and the rest of the protein is
released inside the ER
26
Protein Targeting and Sorting
27
Protein Targeting and Sorting
BiP (binding protein) chaperone in the ER that
binds to hydrophobic regions of the protein
(usually buried in protein structures) BiP is
removed from proteins by ATP hydrolysis, giving
time for the rest of the protein to be
imported and allowed to fold properly so that
the hydrophobic regions are on the
inside protein disulfide isomerase catalyzes
the formation and breakage of disulfide
bonds-- allows various combinations to be formed
to find which gives the most stable
arrangement proteins enter the ER, move from
there to the golgi complex golgi complex
glycosylates proteins for secretion and sorts
them to their correct locations-- default is
to be secreted (via exocytosis) from the
cell specific signals for retention in the ER or
other organelles (ie. lysosomes)
28
Protein Targeting and Sorting
29
Protein Targeting and Sorting
membrane proteins are synthesized very much like
soluble proteins membrane proteins contain the
same ER signal sequence but also have their
own transmembrane sequences for insertion into
the lipid bilayer stop-transfer sequence
hydrophobic a helix which remains in the
membrane transmembrane proteins that have
multiple transmembrane helices have internal
sequences to direct synthesis of each new helix
into the membrane transmembrane helices stay
in the membrane and gets transported to the
golgi in a vesicle while it stays in the membrane
30
Protein Targeting and Sorting
31
Protein Targeting and Sorting
proteins destined for the nucleus, ER,
chloroplasts, and peroxisomes do not go
through the ER but are imported after
translation ie. these proteins are made on free
ribosomes in the cytoplasm special sequences are
used for both the peroxisomes and
nucleus nuclear import sequences sequences
recognized by nuclear pores to allow entry
into the nucleus (ie. polymerases, transcription
factors, etc) most proteins in chloroplasts and
mitochondria are made in the cytoplasm even
though both chloroplasts and mitochondria contain
DNA and their own ribosomes for some of their
own proteins transit sequence N terminal
protein sequence directing cytoplasmic
proteins to the mitochondria or chloroplast,
which is then cleaved off
32
Protein Targeting and Sorting
transit peptidase protease which cleaves off the
transit sequence in the organelle-- can
function very soon after protein enters the
organelle transit sequences are recognized by
specialized transport complexes 2 transit
complexes each are found in mitochondria and
chloroplasts TOM outer mitochondrial membrane
transport complex TIM inner mitochondrial
membrane transport complex TOC outer
chloroplast membrane transport complex TIC
inner chloroplast membrane transport complex to
get to the inner mitochondrial or chloroplast
membranes, transit sequences typically go
directly from TOM to TIM or TOC to TIC electron
microscopy shows 2 membranes of mitochondria
juxtaposed
33
Protein Targeting and Sorting
for proteins entering mitochondria and
chloroplasts, proteins must unfold so they can
fit through the transit pores use chaperone
proteins to keep newly synthesized proteins from
folding completely transit sequence is
recognized, and the protein starts into the
transit pore chaperones outside the organelle
release the chain (requiring ATP) and, after
import, chaperones inside the organelle bind the
transited protein
34
Protein Targeting and Sorting
mitochondria and chloroplasts both have internal
compartments because of their internal
membranes proteins require additional sequences,
hydrophobic sorting signals, to reach their
correct compartment to stay in the membrane,
they require a similar hydrophobic region found
during endoplasmic reticulum protein import and
stop-transfer sequence in general, new
polypeptide chains are recognized as they come
off the ribosome chaperones bind partially
folded proteins so they can be fully
synthesized translocons and transit pores move
chains across the membrane signal sequences are
cleaved off after proteins are correctly targeted