Gene Expression Classification - PowerPoint PPT Presentation
Gene Expression Classification
Gene Expression Classification – PowerPoint PPT presentation
Title: Gene Expression Classification
1
Gene Expression Classification
Y.A. Qi, T.P. Minka, R.W. Picard, and Z.
Ghahramani
Genes
Test error rate
Test error rate
Subjects
Features
Features
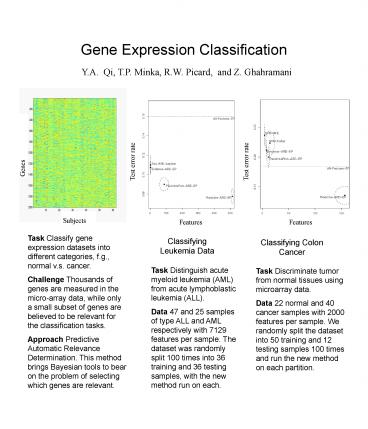
Task Classify gene expression datasets into
different categories, f.g., normal v.s.
cancer. Challenge Thousands of genes are measured
in the micro-array data, while only a small
subset of genes are believed to be relevant for
the classification tasks. Approach Predictive
Automatic Relevance Determination. This method
brings Bayesian tools to bear on the problem of
selecting which genes are relevant.
Classifying Leukemia Data
Classifying Colon Cancer
Task Distinguish acute myeloid leukemia (AML)
from acute lymphoblastic leukemia (ALL). Data 47
and 25 samples of type ALL and AML respectively
with 7129 features per sample. The dataset was
randomly split 100 times into 36 training and 36
testing samples, with the new method run on each.
Task Discriminate tumor from normal tissues using
microarray data. Data 22 normal and 40 cancer
samples with 2000 features per sample. We
randomly split the dataset into 50 training and
12 testing samples 100 times and run the new
method on each partition.
PowerShow.com is a leading presentation sharing website. It has millions of presentations already uploaded and available with 1,000s more being uploaded by its users every day. Whatever your area of interest, here you’ll be able to find and view presentations you’ll love and possibly download. And, best of all, it is completely free and easy to use.
You might even have a presentation you’d like to share with others. If so, just upload it to PowerShow.com. We’ll convert it to an HTML5 slideshow that includes all the media types you’ve already added: audio, video, music, pictures, animations and transition effects. Then you can share it with your target audience as well as PowerShow.com’s millions of monthly visitors. And, again, it’s all free.
About the Developers
PowerShow.com is brought to you by CrystalGraphics, the award-winning developer and market-leading publisher of rich-media enhancement products for presentations. Our product offerings include millions of PowerPoint templates, diagrams, animated 3D characters and more.