the phage lambda (l) - PowerPoint PPT Presentation
1 / 65
Title:
the phage lambda (l)
Description:
... which is used to inject the genetic material into the host. ... Most phages take over the machinery of the host cell to produce a large number of viral particles. – PowerPoint PPT presentation
Number of Views:297
Avg rating:3.0/5.0
Title: the phage lambda (l)
1
the phage lambda (l)
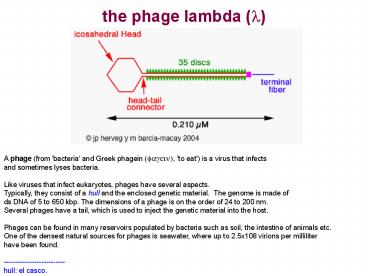
A phage (from 'bacteria' and Greek phagein
(fagein), 'to eat') is a virus that infectsand
sometimes lyses bacteria. Like viruses that
infect eukaryotes, phages have several
aspects.Typically, they consist of a hull and
the enclosed genetic material. The genome is
made ofds DNA of 5 to 650 kbp. The dimensions of
a phage is on the order of 24 to 200 nm. Several
phages have a tail, which is used to inject the
genetic material into the host. Phages can be
found in many reservoirs populated by bacteria
such as soil, the intestine of animals etc. One
of the densest natural sources for phages is
seawater, where up to 2.5x108 virions per
milliliter have been found. ---------------------
----- hull el casco.
2
- Lysis and Lysogeny
- The lambda phage is adorbed on the lamb
receptor. This receptor is only expressed when - the bacteria are grown in the presence of
maltose. Such a bacteriuml is known as
competent. - Lysis multiplication.Most phages take over the
machinery of the host cell to produce a large
number - of viral particles. This result in the lysis of
the host cell, which releases the newborn phages. - Lysogeny rest.
- In some infections, a phage stays silent within
the cell. - This silent phage can go lytic under certain
conditions. - a lysogenic phage is called a prophage
(integrated into the chromosome) - Lysogenic bacteria have immunity against
further infection - The prophage can be induced and is excised from
the bacterial genome
3
Lysis and lysogeny are controlled by the proteins
encoded by cro and cI genes. The lambda phage
will remain in the lysogenic state if cI proteins
predominate, but will be transformed into the
lytic cycle if Cro proteins predominate cI
genes transcription and translation regulate
lysis/lysogeny
4
The cI dimer may bind to any of three operators,
OR1, OR2, and OR3, in the order OR1 gt OR2 gt
OR3. Binding of a cI dimer to OR1 enhances
binding of a second cI dimer to OR2 thus, OR1
and OR2 are almost always simultaneously occupied
by cI dimer . This does not increase the
affinity between cI dimer and OR3, which will be
occupied only when the cI dimer concentration is
high. (a) In the absence of cI proteins, the cro
gene may be transcribed ---gt
lysis (b) In the presence of cI proteins, only
the cI gene may be transcribed
---gtlysogeny (c) At high concentration of cI,
transcriptions of both genes are repressed ---gt
lysogeny When the host DNA is damaged (e.g.,
under UV irradiation), the cI protein may be
cleaved by certain protease promoted by the RecA
protein. Cleaved cI proteins cannot bind to the
operators. Thus, the Cro proteins can be
produced to transform the lambda phage into the
lytic cycle.
5
Lytic or Virulent Phages
6
(No Transcript)
7
(No Transcript)
8
(No Transcript)
9
the phage genome
10
modes of replication Cairn and rolling circle
11
Lysogenic and integration into the bacterial
genome.
- DefinitionA lysogenic phage is a phage that
enters a quiescent state in the cell. - In this quiescent only the cI gene is
transcribed the phage genome exists in a
repressed state. - In this repressed state, the phage DNA is called
a prophage - A prophage dont have the potential to produce
other phages. - In most cases the phage DNA actually integrates
into the host chromosome and is replicated along - with the host chromosome and passed on to the
daughter cells. - The cell harboring a prophage is not adversely
affected by the presence of the prophage and the
lysogenic - state may persist indefinitely. The cell
harboring a prophage is termed a lysogen.
12
2. Events Leading to Lysogeny - The Prototype
Phage Lambda a. Circularization of the
phage chromosome - Lambda DNA is a double
stranded linear molecule with small single
stranded regions at the 5' ends. These single
stranded ends are complementary (cohesive ends)
so that they can base pair and produce a circular
molecule. In the cell the free ends of the circle
can be ligated to form a covalently closed circle
as illustrated below.
13
b. Site-specific recombination In phage lambda,
the integration site is known as att P, in E.
coli the site is att B. The att sites contain
the binding sites for the proteins that mediate
lambda recombination. The integration reaction
(att B x att P) is mediated by the
proteins integrase (Int) and host integration
factor (IHF).
14
integration When integration occurs, two new
sites are created, att L and att R, flanking the
integrated prophage, with no loss of DNA
sequence. All the att sites are alike in that
they contain a 15-bp recognition sequence for the
recombinase (integrase).
excision When att L x att R recombine (mediated
by the proteins integrase and host integration
factor and excisionase Xis), the lambda -DNA is
excised from the E. coli genome, recreating
the att B site in E. coli and the att P site in
lambda.
15
c. Repression of the phage genome - A phage
coded protein, a repressor, is made which
binds to a particular site on the phage DNA,
called the operator, and shuts off transcription
of most phage genes EXCEPT the repressor
gene. The result is a stable repressed phage
genome which is integrated into the host
chromosome. Each temperate phage will only
repress its own DNA and not that from other
phage, so that repression is very specific
(immunity to superinfection with the same
phage).http//pathmicro.med.sc.edu/mayer/phage.h
tm d. Lysogenic conversion - When a cell
becomes lysogenized, occasionally extra
genes carried by the phage get expressed in the
cell. These genes can change the properties of
the bacterial cell. This process is called
lysogenic or phage conversion. This can be of
significance clinically. e.g. Lysogenic phages
have been shown to carry genes that can modify
the Salmonella O antigen, which is one of the
major antigens to which the immune response is
directed. Toxin production by Corynebacterium
diphtheriae is mediated by a gene carried by a
phage. Only those strain that have been converted
by lysogeny are pathogenic
16
Gatway system (Invitrogen)This reaction
involves an entry clone, a destination vector,
and a mix which contains the recombination protein
s necessary for excision and incision (incubation
time one hour).
17
(No Transcript)
18
(No Transcript)
19
The Lambda EMBL3 vector
The Lambda EMBL3 vector (Stratagen, Promega) is a
genomic replacement lambda phage vector capable
of accepting BamH I-compatible fragments (Sau3A
I, Mbo I, Bgl II,or BamH I), ranging in
size from 9 to 23 kb (Figure above). The arms are
prepared by double digestion with Bam H I and
EcoR I followed by a selective precipitation
which removes the small BamH I/EcoR I linker
thatseparate the arms from the stuffer
fragment. Because this treatment leavesthe arms
with BamH I ends and the stuffer fragment with
EcoR I ends, there is no need to physically
separate them. Target DNA cloned into theBamH I
sites of EMBL3 may be removed by digestion with
Sal I.Thered and gam genes are located on the
stuffer fragment therefore, WT phagecannot grow
on the P2 lysogenic strain. When the stuffer
fragment is replaced by an insert, the
recombinant phage becomes red/gam, and the
phage is ableto grow on the P2 lysogenic
strain. Therefore, by plating the library on P2
lysogenic strain, only recombinant phages are
allowed to grow.
20
packaging the phage recombinant DNA
Here are the genes coding for the head and the
tail of the phageProtein coded by the gene
AThe protein A is necessary to fill the head
with one genome and to cut the concatemer
when one genome is packaged into the head.
Without A, the heads stay empty. Protein coded
by the gene E Without E there is no
heads. Protein E is one of the main constituent
of the phage head.
21
(the method described below is only an example,
there other methods)The packaging uses two
kinds of bacteria, each infected by a lysogenic
phages the first phage has a mutation in
protein A in its lytic form, it is unable to
package DNA into a head and to cut the
concatemer at the cos sites the second phage has
a mutation in protein E in its lytic form ,it
doesnt buit a head. the head parts are
disseminated through the cytoplasm. Both phage
are maintained in the lysogenic form by a
thermosensible cI (the protéin acts at
32.Above, it is destroyed and the phage becomes
lytic. The two kind of bacteria show another
peculiarity. Their DNA have not attB
sequences.This mutation doesnt allow this DNA
to be packaged.An extract of both phages is mix
with the recombinant ADN at 45 C. The recovery
is low, between 0.05 and 0.5 .The method can
be used with cosmid DNA.
22
(No Transcript)
23
(No Transcript)
24
(No Transcript)
25
(No Transcript)
26
(No Transcript)
27
(No Transcript)
28
(No Transcript)
29
(No Transcript)
30
(No Transcript)
31
(No Transcript)
32
(No Transcript)
33
(No Transcript)
34
(No Transcript)
35
Adeno-associated-viruses (AAVs)
The 5 serotypes of the human AAVs belong to the
genus Dependovirus.AAVs are similar to
parvoviruses.They are not replicated by any
cells. To replicate them, the host cell has also
to be infected by helpers viruses. The sytem
described above is sold by Stratagen. It is not
dangerous for the people in the lab. It contains
no viruses but 3 plasmids (presented above) an
eukaryotic cell able to produce them (AVV-293
cell), a modified HEK-293 cell.
36
A no helper virus system (eliminating the
requirement for live helpervirus )Stratagenes
AAV Helper-Free System allows the production of
infectious recombinant hAAV-2 virions without
the use of any helper virus.This system takes
advantage of the identification of the 4
specific adenovirus genes (e2A, e4, and va RNA)
that mediateAAV replication and the
demonstration that some of these gene can be
introduced into a modified host cell (AAV-293) by
transfection through the helper plasmid 3 out
of 4 adenovirus gene whose products are required
for the production of infective AAV particles are
supplied on the plasmid pHelper (i.e. e2A, e4,
and va RNA genes). this plasmid is co-transfected
into cells. The remaining adenoviral gene (e1) is
supplied by the AAV-293 host cells. A plasmid to
get more room for the insertthis plasmid bears
the genes re and cap.AAV-293cellsare
HEK293-derived cells with improved
adeno-associated virus production capabilities.
37
ITR inverted repeat sequences (secuencias
repetitivas inversadas)Not I site for the
restriction enzyme Not 1. pAAV-MCS (Multiple
Cloning Site Region, nt 1319-1394)Cla I EcoR
I BamH I Xba IHinc II Hind III
Xho I Bgl
II atcgattgaattccccggggatcctctagagtcgacctgcagaagct
tgcctcgagcagcgctgctcgagagatct
38
(No Transcript)
39
pAAV-MCS (Multiple Cloning Site Region, nt
1319-1394)Cla I EcoR I BamH I Xba
IHinc II Hind III Xho I
Bgl II atcgattgaattccccggggatcctctagagtc
gacctgcagaagcttgcctcgagcagcgctgctcgagagatct
40
rep and cap genes are on another plasmid creating
a 3 kb extra space. Rep and Cap proteins are
implicated in replication and capside
formation respectively.El genóma salvaje AAV-2
contiene los genes rep y cap que codifican Rep y
CAP,proteînas implicadas respectivamente en la
replicaciôn y en la formaciôn de la câpside.
Esos genes contienen dos secuencias terminales
repetitivas (ITRs) que contienen loselementos
cis necesarios para la replicaciôn y el
empaquetamiento.En el sistema sin helper
(helper free), se han metido rep y cap en un
plâsmido. Al sacar êsos genes, se crea un
espacio de 3kb que son los utilizados para ahî
meter el inserto.
41
The helper plamid contains 3 genes that force
the host cell to replicate the virus producing
particles able to infect the target cell.El
plásmido "Helper que contiene tres de los genes
(E2A, E4 y VA RNA) que obligan a la célula
huésped a replicar el virus y a producir
partículas capáces de infectar la célula target.
42
4. Las cêlulas huêsped, AVV-293 Esas cêlulas
pueden proveer a otros genes virales,
notablemente el gen E1 de los adenovirus.
43
examples
Human immunoglobulin inhibits liver
transduction by AAV vectors at low AAV
neutralizing titers in SCID mice Ciaran D.
Scallan, Haiyan Jiang, Tongyao Liu, Susannah
Patarroyo-White, Jurg M. Sommer, Shangzhen Zhou,
Linda B. Couto,and Glenn F. PierceBlood, 1
March 2006 Volume 107, Number 5
44
Adeno-associated virus (AAV) vectors have
reproducibly delivered transgenes in animal
models, resulting in successful treatment and
cure of numerous clinically relevant
diseases.These studies have demonstrated
sustained transgene expression without toxicity
in mice, rats, dogs, and nonhuman primates. In
addition, severa lclinical trials using AAV2
vectors have shown excellent safety profiles
however, clear evidence of therapeutic efficacy
has not yet been Achieved. A major challenge for
AAV-mediated genetherapy is pre-existing
AAV-specific antibodies prevalent in the human
population resulting from prior natural
infections. Theeffect of these pre-existing
antibodies on AAV gene transfer variesamong
individuals, depending on actual neutralizing
titers, thedelivery route, and target
organ. Compartments that have limited access to
systemically circulating antibodies, such as
brain and skeletal muscle, have been transduced
successfully in the presenceof high
circulating Neutralizing titers. Intravascular
delivery to target organs such as the liver will
likely be more Problematic because the
circulating antibodies can rapidly bind to
antigens at an average Kd of 10 7 to 10 11M.
45
Adenoviruses
Human serotype 5 adenoviruses are also used to
introduce in mammalian cells, genes we want to
express. Adenoviruses dont need cell in a
reproductive phase. Serotype 5,which is rendered
replication defective by deletion of the genes E1
and E3. This deletion gives an insertion space of
7.5 kb E1 is essential for the assembly of
infectious virus particles and is
complemented E3 encodes proteins involved in
evading host immunity. As E1 is necessary, we
complement it by modified HEK 293 cell that
express E1 (AD-293).As the virus posses a very
small number of restriction sites, the gene of
interest is first inserted in a shuttle vector.
Once the gene is inserted in the shuttle, it is
inserted in pEasy by recombibation
46
Because the adenovirus genome is large (36 kb)
and contains few restriction sites, the gene of
interest is first inserted into a shuttle vector
and then transferred into the adenovirus genome
by means of homologous recombination. Employing
the efficient homologous recombination machinery
in E. coli, a recombinant adenovirus is
producedby a double-recombination event between
cotransformed adenoviral backbone plasmid vector,
pAdEasy-1, and a shuttle vector carrying the gene
of interest. This eliminates the need to
manipulate the large adenovirus DNA molecule in
vitro (in restriction and ligation reactions).
47
ES encapsidatoion site
48
(No Transcript)
49
(No Transcript)
50
(No Transcript)
51
practice
--------------------------- balance (aparato)
balanza sterile petri dishes placas Petri
estériles5 tubes with metal caps 5 tubos de
ensayo con tapas de metal. an "Erlenmeyer" flask
with a gauze cap un Erlenmeyer provisto de
buchón estéril.Gilson pipettes on a white rack
and tips pipetas Gilson y puntas spatula
paleta, espátula
52
- Select the material you'll use today
- Prepare the petri dishes with two agar layers
bottom and top - bottom agar 1,5 g of agar in LB medium (LB
Luria-Bertoni) - top agar a 1/1 dilution of botoom agar
- ---------------------------
- Selecciona el material a usar.
- Prepara placas de agar con dos capas
- 5 sterile petri dishes 5 placas Petri
estériles5 tubes with metal caps 5 tubos de
ensayo con tapas de metal. - Bottom and top agar la capa inferior (agar de
base) y la capa superior.
53
3. Sterilize both agar bottom agar in an
erlenmeyer top agar in tubes.4. Pour Bottom
agar in petri dishes (using the laminary flow
hood)
5. Place top agar tubes at 47 while you are
infecting the competent E. coli.
---------------------------------3. Estériliza
en una olla de presión el agar de base (en
erlenmeyer) y el agar superior (en tubos de
ensayo). 4. Vierte el agar de base en las placas
(en la cámara de flujo laminar) 5. Manten el agar
superior a 47 C mientras infecta la E. coli.
competente
54
6. Mix the infected E coli with top agar, mix,
and pour on top of bottom agar.
7. Let cool down in the incubator now you have
two layers
-------------------------- 6. Siembra la E. coli
infectada (mezclada con el agar superior a
47C)y viertelo sobre el agar de base.7. Dejalo
enfriar antes de meterlo en la estufa.
55
- Select the material you'll use today
- Prepare the petri dishes with two agar layers
bottom and top - bottom agar 1,5 g of agar in LB medium
- top agar a 1/1 dilution of bottom agar
56
3. Sterilize both agar bottom agar in an
erlenmeyer top agar in tubes.
57
4. Pour Bottom agar in petri dishes (using the
laminary flow hood)
58
5. Place top agar tubes at 47 while you are
infecting the competent E. coli.
59
6. Mix the infected E coli with top agar, mix,
and pour on top of bottom agar
60
(No Transcript)
61
(No Transcript)
62
(No Transcript)
63
(No Transcript)
64
(No Transcript)
65
(No Transcript)