Genetics PCB 3063 PowerPoint PPT Presentation
1 / 35
Title: Genetics PCB 3063
1
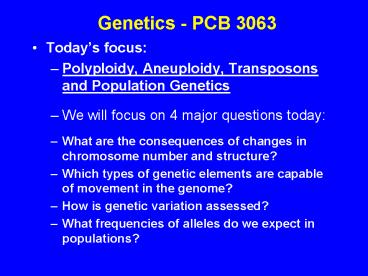
Genetics - PCB 3063
- Todays focus
- Polyploidy, Aneuploidy, Transposons and
Population Genetics - We will focus on 4 major questions today
- What are the consequences of changes in
chromosome number and structure? - Which types of genetic elements are capable of
movement in the genome? - How is genetic variation assessed?
- What frequencies of alleles do we expect in
populations?
2
Polyploidy and Aneuploidy - Terms
- Although diploid organisms are common, not all
organisms are diploids
- Often, monoploid organisms (like our old friend
Neurospora crassa) are called haploid. - Organisms that contain only complete sets of
chromosomes are called euploid. - If an organism does not have one or more complete
sets of chromosomes, it is called aneuploid. - In many cases, aneuploidy is associated with
pathological phenotypes.
3
Some Polyploids are Infertile
- Triploids and other polyploids with odd numbers
of chromosomes are often infertile. - This reflects the fact that the unpaired
chromosomes will segregate randomly. - For example, a triploid organism with 4
chromosomes will give rise to different classes
of gametes, e.g. - The probability of 2N gamete is 0.0625 based upon
the multiplication rule. - In some cases, this infertility can reflect
abnormalities of gametes with incomplete sets of
chromosomes. - This infertility may be limited to male or female
gametes.
4
Tetraploids from Triploids
- Triploids can give rise from tetraploids.
- This is typically called passage through the
triploid block.
2N
2N
2N
1N
Pollen
3N
Pollen Various Chromosome Numbers
4N
Most common viable offspring
5
Types of Polyploids
- Chromosome sets in polyploids can have a number
of distinct origins. - If the chromosome sets are sufficiently similar
that they can pair, usually reflecting fusion of
unreduced (2N) gametes within a species, the
polyploid is called an autopolyploid. - If the chromosome sets are dissimilar and unable
to pair, usually reflecting the fusion of gametes
from different species, the polyploid is called
an allopolyploid. - In some cases, usually when parental species are
known, allotetraploids are called amphidiploids. - Allopolyploids can be of two major types
- If the chromosomes cannot pair at all, they are
often called genomic allopolyploids. - Result from the fusion of gametes from fairly
distinct species. - If the chromosomes pair partially, so there are
regions that cross-over, they are often called
segmental allopolyploids. - Reflect fusion of gametes from relatively closely
related species.
6
Types of Polyploids - Allopolyploids
- Allopolyploidy can allow crosses of relatively
distinct species...
- But sometimes the crosses arent valuable.
- E.g., the Rabbage was an attempt to cross the
radish (2N18) and cabbage (2N18). The cross was
infertile but allowing the chromosome number to
double generated fertile plants. However, rabbage
plants had small roots and small leaves...
7
Plants, Animals and Polyploids
- Polyploidy has played different roles in the
evolution of plants, animals and fungi. - Polyploidy is very common in plants, but rare in
the animals and fungi. - This may reflect the ability of plants to
reproduce vegetatively, at least in part. - The paucity of polyploid fungi is unclear.
- More than 70 of angiosperms and 95 of ferns
have polyploid ancestors. - Both maize and Arabidopsis are diploids but they
show clear evidence of being polyploids that
reverted to disomic inheritance. - In both of these organisms, many genes are
duplicated and the duplications occur in blocks. - For maize, more than 85 of loci appear to have
been duplicated. - See http//www.agron.missouri.edu/mnl/73/47braun.h
tml for examples of a few duplicated maize loci,
if you are interested.
8
Plants, Animals and Polyploids
- Despite the paucity of polyploid animals and
fungi, these processes have played a role in the
evolution of these groups. - S. cerevisiae appears to have been a tetraploid
that reverted to disomic inheritance. - Like Arabidopsis, there are a number of
duplicated blocks in the S. cerevisiae genome. - So the ancestor of S. cerevisiae was probably a
tetraploid that reverted to disomic inhertiance
and lost many duplicate loci. - Vertebrates also have a large number of duplicate
genes. - These have been proposed to reflect two rounds of
duplication due to the reversion of tetraploids
to disomic inheritance. - This idea is still controversial -- the
duplicated genes in vertebrates could simply
reflect a period of active gene duplication.
9
Aneuploidy
- In normal diploids, individuals are said to be
disomic for all chromosomes. - If three copies of a specific chromosome are
present, the individual is said to be trisomic
for that chromosome. - Likewise, if one copy of a specific chromosome
are present, the individual is monosomic for that
chromosome. - Cases in which two chromosomes are present in
unusual numbers are referred to as cases where
the organism is doubly trisomic or monosomic. - With the exception of sex chromosomes, humans
tolerate trisomy and monosomy poorly. - Down syndrome is trisomy 21, and it is associated
with a number of phenotypes. - Trisomics for the larger chromosomes in humans
are lethal during development, and embryos that
have these abnormalities undergo spontaneous
abortion.
10
Aneuploidy
- Thus, most aneuploidies in humans involve the sex
chromosomes - Down syndrome - trisomy 21.
- Turner syndrome - single X chromosome.
- Short stature, limited sexual development, mental
retardation - Klinefelter syndrome - multiple X chromosomes
along with a Y - Males, but some female characteristics.
- XYY - male, no clear phenotype.
- The basis for the viability of these individuals
is likely to be the fact that dosage compensation
mechanisms exist for the sex chromosomes. - Also, the Y has very few genes, so little
potential to disrupt development. - The fact that these syndromes do have phenotypes
indicates that dosage compensation is imperfect.
11
Aneuploidy of Sex Chromosomes
- The potential for viable sex chromosome
aneuploids is also seen in other organisms.
- This shows a bilateral gynandromorph -- an
organism in which a mitotic nondisjunction has
occurred. - Thus, this organism is a somatic aneuploid.
- Since this is a moth, what would you call the sex
chromosomes that were involved?
- At a cellular level, aneuploidy can play a major
role in the development of cancer. - E.g., loss of chromosomes or chromosome regions
with tumor suppressors.
12
Chromosomal Rearrangements
- A number of chromosomal rearrangements are
described in your text. - Rearrangements can lead to pathological
conditions. - Deletions of regions lead to specific phenotypes.
- This has been used to map genes in some organisms
(e.g., Drosophila). - Alternatively, fusion of a strong promoter from
one gene to another gene can cause unusual
phenotypes. - The yellow allele of the mouse agouti gene
described in chapter 4 (page 105) is a deletion
that results in the overexpression of the agouti
gene. The lethality of the allele actually
reflects deletion of a gene adjacent to the
agouti gene that is essential for viability. - Burkitt's lymphoma (BL) is characterized by
translocations between chromosome 8 and 14 (90
of cases). The c-myc protooncogene maps to the
chromosome 8 breakpoint and Ig (immunoglobulin)
genes map to the chromosome 14 breakpoint.
Placing the c-myc gene near the Ig promotor
(active in B lymphocytes) results in increased
expression of c-myc - which activates cellular
proliferation.
13
Chromosomal Rearrangements
- However, it is important to stress that these
rearrangements occur during evolution and
represent on mechanism isolating species. - For example, two species of dik-diks were
recognized until people examined karyotypes,
revealing three species. - The cryptic species of dik-dik cannot hybridize,
indicating that the karyotypic differences are
meaningful.
14
Transposons
- Transposons - jumping genes
- The phenomenon of transposition was originally
noted by Marcus Rhoades and Barbara McClintock in
maize. - Rhoades noted an unusual pattern of reversion at
the A1 locus. - A1 (anthocyaninless1) is necessary for formation
of the purple anthocyanin pigments in maize. - The dominant allele (A1) is colored while the
recessive (a1) is clear. NOTE THAT THE TEXT IS
INCORRECT HERE. - Rhoades noted that a specific a1 mutant actually
reverted multiple times within a kernel of corn -
giving a dotted appearance
Note the spotted kernels - the spots are due to
the transposon jumping out of the A1 gene.
15
McClintock and the Ac-Ds System
- Barbara McClintock noted that recessive markers
on maize chromosome 9 could be revealed at a high
rate in a specific mutant. - This phenotypic change was accompanied by a
cytological change - loss of part of the short
arm of chromosome 9!
16
McClintock and the Ac-Ds System
- McClintock called the breakpoint in these lines
Ds, for dissociation. - The break would not occur unless an Ac element
(activator) was crossed in.
17
What are Ac and Ds?
- Ds elements can also move in the genome.
- Thus, they did not behave in the way genes have
been shown to behave previously - they did not
have defined loci in the genome! - In addition, they relied upon the Ac element for
activity. Why? - When Ac and Ds elements were cloned, it was shown
that they were related. - Ac has two open reading frames (ORFs), while Ds
elements have deletions covering at least part of
the first ORF.
18
What are Ac and Ds?
- When Ac and Ds elements were cloned, it was shown
that they were related. - Ac has two open reading frames (ORFs), while Ds
elements have deletions covering at least part of
the first ORF. - The first ORF is a transposase...
- It is necessary for insertion at novel loci,
excision, and chromosome breakage. - Ac will supply the transposase in trans -
activating the Ds elements.
19
Examining Transposons in Maize
From Feschotte, Jiang and Wessler (2002) Nature
Reviews Genet. 3 329-41.
20
Types of Transposons
- Ac and Ds are DNA transposons.
- The transposition does not involve an RNA
intermediate. - Ac is an example of an autonomous element (can
transpose by itself). - Ds is a non-autonomous element (requires
trans-acting factors from another element). - In this case, Ac supplies the transposase - the
trans-acting factor. - Bacterial insertion sequences and Drosophila P
elements are DNA transposons. - A particularly interesting group of DNA
transposons are the MITEs -- - MITEs predominate in the non-coding regions of
grass genes. - MITEs elements are structurally reminiscent of
non-autonomous DNA transposons, are small (lt 600
bp) and have terminal inverted repeats. However,
their high copy number, target-site preference
(TA or TAA) and the uniformity of related
elements distinguished them from the previously
described transposons.
21
Types of Transposons
- Other transposons are retrotransposons.
- These elements have an RNA intermediate.
- In many cases, they resemble retroviruses but
lack the env (envelope) gene. - Some elements transpose through an RNA
intermediate but are distinct from retroviruses. - These include the AluI elements and L1 elements
of humans. - The human genome has more than 500,000 AluI
elements and 100,000 L1 elements.
22
Assessing Genetic Diversity
- A variety of methods have been used to examine
genetic variation in populations - Observation of phenotypes
- This is inexpensive and straightforward
- Problematic for several reasons
- Some genes contribute to continuous variation.
- Some genes have unknown phenotypes.
- Examination of proteins
- Typically done using electrophoresis.
- Direct examination of DNA variation
- Can be accomplished by sampling a subset of sites
in DNA, either at a specific locus or across the
genome. - Can be accomplished by sequencing loci.
- Sequencing is still relatively expensive.
23
Protein Electrophoresis
- Protein variation is often examined using gel
electrophoresis - Usually, either starch or cellulose acetate gels
are used. - The gels are run under non-denaturing conditions,
so enzymes can be detected by stained with
substrates that change color. - Migration is determined by charge and size of the
proteins - most variation is charge. - Many proteins will migrate to the anode () but
the same principle works for the cathode as well
24
How do Variants Migrate?
- Imagine a population that has two different
alleles of a gene encoding a protein we are
examining - If the alleles encode proteins with different
charges, the are likely to migrate at different
rates. - Typically, these variants are called fast and
slow. If so, we will see - If the enzyme is not a monomer, more complex
patterns will result. - For example, a dimer would yield
- What are the bands composed in this gel?
A - slow homozygote. B - heterozygote. C - fast
homzygote.
A
B
C
A
B
C
A - slow homozygote. B - heterozygote. C - fast
homzygote.
25
RFLP - Examining DNA Variation
- RFLPs are Restriction Fragment Length
Polymorphisms. - Restriction enzymes cut at defined sequences in
DNA. For example, EcoRI cuts at - If an EcoRI site is present in some members of a
population but not others, it is an RFLP. - Alternatively, the polymorphism my reflect a
large indel between two EcoRI sites. - For a single locus, one can detect RFLPs using
either PCR or Southern blotting. - For a Southern, DNA is sized (using agarose gel
electrophoresis) - Then the DNA is transferred to a membrane.
- Labeled probe DNA (corresponding to the locus of
interest) is hybridized to the membrane. - Alternatively, one PCR amplifies the locus and
cuts with the enzyme of interest.
----GAATTC---- ----CTTAAC----
26
Multilocus (RFLP) Fingerprinting
- However, a much more powerful approach was
proposed by Alec Jeffries in the 80s. - This is multilocus RFLP analysis, often called
DNA fingerprinting. - One uses a probe to VNTR (variable number tandem
repeat) sequences and then examines a Southern
blot of DNA cut with various enzymes. - Usually the repeat number changes sufficiently
rapidly that most populations will show
differences among individuals. - This method is commonly used in forensics and for
paternity analysis.
A DNA fingerprint blot
27
PCR
- In contrast, PCR methods are limited to known
loci. - This reflects the need for primer sequences.
- PCR is popular because little template DNA is
necessary. - Most of the template in later rounds was
generated in early rounds.
28
RAPD PCRExamining Multiple Loci
- Because so little DNA is necessary, PCR is
desirable for population surveys. - Likewise, PCR is very useful for forensics.
- Can it be used to survey multiple loci?
- One method is to use short primers with arbitrary
sequences, but anneal at a lower temperature. - Typically, one uses specific primers that are 18
to 30 mers and anneals at temperatures over 50C. - Under these conditions, typically only one locus
will anneal to both primers and amplify. - However, RAPD PCR uses shorter primers (12 mers
in some cases) and lower temperatures. - Many loci in the genome will anneal to these
primers and amplify.
29
RAPD PCR Related Methods
- RAPDs still requires regions that match the
primers. - But the low annealing temperature and short
sequence length allow the primers to anneal at
many more places than in a standard (locus
specific) PCR reaction. - Since the specificity is so low, one actually
uses a single primers. - This will amplify only loci that have two
flanking sites that can anneal to the primer. - Do you think the loci amplified by RAPD PCR will
correspond to known loci? or to protein coding
loci? - Can you distinguish homozygotes from
heterozygotes using RAPDs? - A number of similar techniques have been
developed (e.g., ISSRs) as have methods that use
PCR to scan polymorphic restriction sites (AFLPs).
30
SNPs - Sequence Variation
- One can also amplify specific loci by PCR and
examine the sequences. - Nucleotide sites that are polymorphic are called
single nucleotide polymorphisms or SNPs. - This method is a much more powerful method to
examine all variation at a specific locus. - However, it is expensive.
- It can only examine a single locus at a time.
- Sequencing can detect heterozygotes.
- Heterozygotes will show both peaks in a
sequencing reaction.
31
Microsatellites - Fragment Analysis
- Like VNTRs (minisatellites) used in DNA
fingerprinting, other types of repeat loci are
popular targets in population studies. - Microsatellites are repeats of 2 or 3 nucleotides
(like ...GTGTGTGTGTGTGTGT) - Microsatellites often vary in length due to
slippage of DNA polymerase during replication.
32
Microsatellites - Fragment Analysis
- Microsatellite repeat loci are popular targets in
population studies. - If one designs primers to a specific
microsatellite locus (in the flanking
non-repetitive region) one can amplify the
microsatellite by PCR. - Then, one can separate the fragments by size
using a DNA sequencer to look for size
polymorphisms. (dont actually sequence)
33
What do we do with this data?
- Information about genetic variation in not very
useful in the absence of methods to analyze the
data. - Typically one examines specific patterns that one
expects for the variation. - The simplest expectation is given by
Hardy-Weinberg equilibrium. - If we have a population in which individuals are
randomly mating and there are two alleles at a
locus, the expected proportions of homozygotes
and heterozygotes is - We expect p2 homozygotes for allele 1, where p is
the frequency of allele 1. - Likewise, we expect q2 homozygotes for allele 2,
where q is the allele 2 frequency. - Finally, we expect 2pq heterozygotes, where p and
q are defined above.
34
What do we do with this data?
- The simplest expectation is given by
Hardy-Weinberg equilibrium. - If we have a population in which individuals are
randomly mating and there are two alleles at a
locus, the expected proportions of homozygotes
and heterozygotes is - We expect p2 homozygotes for allele 1, where p is
the frequency of allele 1. - Likewise, we expect q2 homozygotes for allele 2,
where q is the allele 2 frequency. - Finally, we expect 2pq heterozygotes, where p and
q are defined above.
It is easy to think of Hardy-Weinberg equilibrium
graphically, like this...
35
TestingHardy-Weinberg Equilibrium
- How do we test whether the data we have collected
fit Hardy-Weinberg equilibrium? - One way is to use our old friend, the c2 test!