RNA Splicing PowerPoint PPT Presentation
1 / 20
Title: RNA Splicing
1
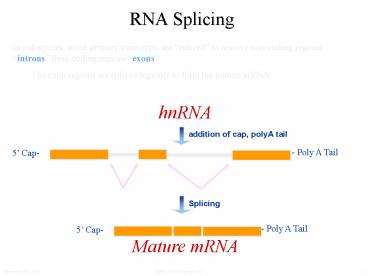
RNA Splicing
In eukaryotes, some primary transcripts are
spliced to remove non-coding regions introns
from coding regions exons The exon regions are
spliced together to form the mature mRNA
hnRNA
addition of cap, polyA tail
- Poly A Tail
5 Cap-
Splicing
- Poly A Tail
5 Cap-
Mature mRNA
2
Types of mRNA Splicing
Types I II self-splicing of catalytic RNA
sequences (Ribozymes) - Two types reflect
different ribozyme types Type III occurs in a
protein - RNA complex-the Splicesome - It appears
that RNA components of this structure largely (if
not exclusively) provide the catalytic functions
i.e. they act as ribozymes
3
Prevalence Of Splicing
Type III
- most mRNAs in vertebrates - many mRNAs in
invertebrates - some mRNAs in unicellular
eukaryotes
mRNA Eukaryotes
no type III mRNA splicing in Prokaryotes!
Type I II
Eukaryotes
- rRNA in tetrahymena nuclei (Type I) - mRNA
rRNA in fungal mitochondria (I II) - mRNA in
some chloroplasts (II)
Prokaryotes
- mRNA in bacteriophage T4 (I)
4
Mechanism of pre-mRNA Splicing(Spliceosome
Mediated) Type III
- - Introns have conserved sequences at the splice
junctions - - SnRNPs (Small nuclear ribonucleoprotein
particles) bind critical sites on the pre-mRNA - - Acronym is pronounced SNURPs
- - these are complexes containing both protein and
small RNAs - the small RNAs are transcribed by RNA
polymerases II and III - they then associate with accessory proteins
- the complex then recognizes critical sites for
splicing by base pairing
5
Once all of the different snRNPs associate with
their appropriate targets on the pre-mRNA ? the
entire (very large) complex is called a
Spliceosome Types of snRNPs U1 binds 5
splice site U2 binds Branch point and exposes
key A nucleotide U5 binds 3 splice site U4
U6 also required function less well defined
6
Table 28.6
- consensus sequences are conserved throughout
eukaryotes
Conservation of sequence is expected, since
recognition of sequences is accomplished by base
pairing with snRNPs RNA component
7
Figure 28.31 of Mathews
Secondary structure model of human U1 snRNP. The
region where it recognizes the pre-mRNA is also
shown
8
Figure 28.32 of Mathews Schematic view of the
proposed mechanism of splicing Exons in red
(dotted) Introns are black The products are a
spliced message and a lariat which is then
degraded
2OH of A reacts with G at 5 end of
intron-called trans-esterification reaction
Second trans-esterification reaction
Lariat
Unusual 2 5 linkage
9
Lariat product of RNA splicing reaction.
A from branch site and G from 5 end of
intron are connected via a 2 5 Linkage
10
Figure 28.33 of Mathews Proposed model for the
mechanism of Type III splicing
lariat
11
Self-Splicing (Type I)
T.R. Cech, 1982 made a revolutionary discovery
RNA could have catalytic activity RNA
enzyme Ribozyme (look at pages 395-397 in
Mathews)
- This type of splicing requires only
- Pre-rRNA
- Mg2
- G (GDP,GMP, guanosine) guanine does not work
12
- Self-Splicing Introns fold so as to
- Bring 5 3 splice sites together
- Form a G-binding pocket close to the splice sites
- Facilitate the action of Mg2, which
- Stabilizes the folded RNA structure
- Undoubtedly helps to lower ?G? for bond transfer
- 3-OH of the G-containing cofactor initiates the
reaction - - attacks phosphate at the 5 splice site.
- - itself becomes attached to the 5 end of the
intron - - Role equivalent to branch site adenine in type
III splicing
13
Figure 11.30 of Mathews Type I splicing
Steps 1. Looping of IVS 2. Attack by pGOH 3.
Cleavage at 5 end of intron 4. 3 U attacks 3
end of IVS 5. rRNA is spliced and IVS is released
14
Alternate Splicing
Splicing at alternate splice sites can lead to
the production of several gene products from one
gene - Proteins involved in splicing can bring
alternate domains of pre-mRNA together to give
alternate splicing events - Alternate splicing
can be regulated - differential splicing can
occur at different times during development and
in different cell types
GENE (hnRNA)
2
3
4
1
1
2
3
4
2 Different mRNA products
1
3
4
15
Figure 28.34 Example of alternate splicing with
rat ?-tropomyosin
16
Splicing can be regulated by repressors and
activators
17
Splicing enhancers are sequences within RNA
transcripts that can promote alternative
splicing. They bind regulatory proteins that can
act as either activators capable of recruiting
key splicing factors to weak splice sites
(those sites that can be overlooked by splicing
machinery if enhancers are not present), or
repressors that can interfere with the inclusion
of a particular exon
18
In addition to alternative splicing, the use of
alternative promoters also generates diversity in
patterns of gene expression In this example, the
same protein is encoded, but the 5 UTR regions
are different therefore, the transcript may have
different properties and be regulated in
different ways e.g. there might be different
translation signals in the 5 UTRs that could
promote or inhibit translation.
Translation initiation site
19
An extreme example of how splicing, plus other
types of processing, can give rise to two
completely different gene products from the same
gene - the calcitonin/neuropeptide gene in
thyroid cells versus neurons.
1
2
3
20
Summary of general gene organization and
processing