Protein Synthesis PowerPoint PPT Presentation
1 / 122
Title: Protein Synthesis
1
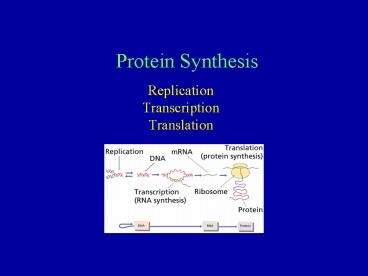
Protein Synthesis
- Replication
- Transcription
- Translation
2
?
3
Histones
4
How is Heritable Information Transferred?
- Griffith, 1912
- Science as a Process
5
What Is the Genetic Material
- Thought to be proteins
- 20 amino acids can create greater variety
- DNA has only different bases
- A, T, C, G
6
Role of DNA in Inheritance
- Griffith had two strains of Streptococcus
pneumoniae, one harmless the other pathogenic - Parts of heat-killed virulent strain were mixed
with harmless strain - Harmless strain became pathogenic
7
Heat-killed S-strain R-strain
Heat-killed S-strain
R-strain
S-strain
8
(No Transcript)
9
?
10
Avery MacLeod - McCarty
- 1944 DNA is the transforming
molecule
"Studies on the Chemical Nature of the Substance
Inducing Transformation
of
Pneumococcal Types Induction of
Transformation by a
Desoxyribonucleic Acid Fraction
Isolated from
Pneumococcus
Type III"
Oswald Avery
11
(No Transcript)
12
Hershey-Chase
- Science as a process 1952
- Used Bacteriophages (virus)
- Viruses are DNA or RNA
- Invade bacteria
- Nucleic acid surrounded by a protein capsule
Martha Chase, Alfred Hershey
13
Is protein the genetic material? No radioactive
S35 in the pellet (bacteria) therefore genetic
material is NOT protein the capsule didnt cause
genetic change
14
Is DNA the genetic material? Grow phages with
P32 when phages invade bacteria, bacterial DNA
becomes radioactive P32 found in bacterial
pellet
15
Why?
Hershey-Chase Blender
- In testing protein why grow the
phages in S35? - Because proteins have SH bonds that DNA does not
have - In testing DNA why grow the phages in P32?
- Because DNA has phosphate groups
that proteins dont have
16
(No Transcript)
17
Structure of DNA
- Sugar (deoxyribose)
- Phosphate group
- Nitrogenous base
18
Four nitrogenous bases uracil (RNA) Purines
adenine, guanine (9-sided) Pyrimidines
cytosine, thymine
19
Discovery of Structure of DNA
- Rosalind Franklin 1952 X-ray crystallography
(scattering of X-ray by reflection with
particles) - Suggested sugar-phosphate backbone
20
(No Transcript)
21
(No Transcript)
22
(No Transcript)
23
Discovery of Structure of DNA
- 1953 Watson-Crick wrote paper on
double helix model received Nobel
Prize in 1962
24
(No Transcript)
25
DNA Replication
- Base pairing enables existing strands to be
unwound - Unwound strand acts as a template for a new strand
DNA replication
26
Semi-conservative Model
- Current model
- Two new stands
- Half from each old strand
27
Meselson-Stahl Experiment
- Model of DNA replication
- 1950s
- Scientific process
28
(No Transcript)
29
(No Transcript)
30
(No Transcript)
31
(No Transcript)
32
DNA Replication
- Origin of replication
- Specific sequence
of nucleotides - Replication fork
both directions at the
same time - Replication bubble
33
Elongating A Strand
- Nucleoside
tri-phosphates - Adenosine
tri-phosphate, etc. - Two phosphate
groups are
dropped - Enzyme DNA POLYMERASE
34
2 strands of DNA are anti-parallel 3 and 5
carbon atoms nucleotides can ONLY be added to
the 3 end elongation can only occur in the 5
to 3 direction
35
2 strands leading and lagging Leading strand
(adding nucleotides 5 to 3) is continuous
Lagging is in fragments
36
DNA Replication
- 2 strands leading, lagging
- Leading strand adds to the 3 end and is
continuous - Lagging strand - nucleotides are added in
fragments
37
(No Transcript)
38
DNA Elongation
- DNA must be primed
- Nucleotides can only be added to an new strand
that is already started - Primers are segments (10 nucleotides) of RNA
- 3 to 5 additions are done continuously so
leading strand is only primed once - Lagging strand has to be primed constantly
- DNA ligase then glues Okazaki fragments together
39
DNA Elongation
- Primase enzyme that binds primers to 3end of
the DNA - DNA Polymerase enzyme that adds nucleotides to
template - Ligase enzyme that glues fragments together
40
Lagging strand
41
Goofs
- Mismatch repair special proteins that remove
and replace incorrectly sequenced nucleotides - Colon cancer
- 100s of repair enzymes
prevent goofs from becoming
part of the genome - goofs may be a cause of ?
- Evolution
42
The Ends Problem
- Primer at the 5 end is
left after replication
(not replaced with DNA) - Nucleotides only added
at 3 end - 5 end is lost
(not copied next time)
43
The Ends Problem
- Prokaryotes have circular chromosome
- No ends to be lost
- Eukaryotes have Telomeres
- Telomere multiple repeats of
nonsense DNA (no genes) - 100s 1000s of nucleotides
44
The Ends Problem
- Telomeres also prevent cell from recognizing
damage to the DNA and apoptosis - Telomerase enzyme that
lengthens DNA, replace lost
nucleotides - Lack of may cause aging (?)
- Cancer too much telomerase (?)
45
Genes To Proteins Chapter 17
- Transcription and Translation
46
What is a Gene?
- Introns, leaders, trailers
- RNAs
- One gene one polypeptide theory
- Expressed proteins are the organisms phenotype
47
Gene to Protein
- Transcription
- Copy gene on DNA into mRNA
- Translation
- Decode transcribed mRNA into primary structure of
protein
48
Transcription
- DNA copied into mRNA pre-RNA
- RNA is processed into finished RNA
49
(No Transcript)
50
(No Transcript)
51
(No Transcript)
52
(No Transcript)
53
Gene to Protein
- 4 nucleotide bases
- 20 amino acids (8 essential)
- Combinations of 3 nucleotides produces the code
for a specific amino acid triplet code - Codon
54
(No Transcript)
55
Codons Decoded
- 1960s Nirenberg
- Added poly UUU to amino acids/ribosomes/etc.
- Produced amino acid phenylalanine
- UUU must code for phenylalanine
56
Dictionary of genetic code
AAA, CCC, GGC, UAA, AUG, UCA, ACC, ACU
57
Genetic Code
- Redundancy more than one codon for some amino
acids - No ambiguity codons only code for one specific
amino acid
58
Genetic Code
- Codon is the SAME for every organism bacteria to
humans - UNITY WITHIN DIVERSITY
- EVOLUTION similar codons/amino acids indicate
common ancestry
59
Transgenic organisms
60
Transcription
- Synthesis and Processing of RNA
- Initiation
- Elongation
- Termination
61
Protein Synthesis
- Transcription
- Translation
62
Initiation
- RNA polymerase unzips DNA
- Attaches RNA nucleotides to complimentary bases
of DNA - Reads DNA 3 to 5
- RNA read 5 to 3
63
RNA
- RNA nucleotides use ribose as sugar
- Replace thymine with uracil
- A-U
64
(No Transcript)
65
Initiation
- Promoter sequence of DNA where the RNA
polymerase attaches - Starting point
- Which side of the two strands is read
66
Promoter
- TATA box - short sequence of thymine/adenine
- Upstream of promoter
- Start here message
- Transcription factors (proteins) attach to TATA
box - RNA polymerase attaches and reads DNA
67
Promoter
- Gene expression is controlled by a variety of
factors - Transcription protein factors
- TATA box
68
Elongation
- RNA polymerase reads about 30-60 nucleotides per
second - Several RNA polymerases can transcribe together
lots of mRNA - DNA reforms behind RNA polymerase
69
Termination
- Termination site on DNA
- Prokaryotic RNA is ready to be translated
- Eukaryotic RNA must be modified and carried to
ribosomes for translation
70
(No Transcript)
71
(No Transcript)
72
(No Transcript)
73
(No Transcript)
74
Eukaryotic Cell mRNA Processing
- 5 and 3 ends are modified
- Cap at the 5 end
- Protects growing strand from hydrolytic enzymes
- Helps ribosome to recognize attachment site to
begin translation - GTP leader (noncoding sequence)
75
- 3 has a trailer and a poly A tail attached
- Trailer noncoding sequence
- Poly A tail long chain of adenine nucleotides
- Prevents degeneration of RNA
- tags mRNA for leaving the nucleus
- Added after the stop codon
76
mRNA Splicing
- Much of DNA is non-coding
- mRNA copies all parts (non-coding)
- Heterogenous Nuclear RNA unfinished, raw
- hnRNA Pre-RNA
- Non coding sequences are removed before leaving
the nucleus
77
- Introns sequences that do not contain protein
codes excised from pre-RNA - Exons sequences of RNA that are translated
- RNA Splicing introns removed and exons
reattached together - Ribozymes RNA that functions as an enzyme
78
RNA Splicing
- Small, nuclear ribonucleoproteins (snRNPs)
snurps packages of protein and RNA found
only in the nucleus - Recognition sites for attachment of ribozymes
79
- snRNPs form Spliceosomes
- Spliceosome binds the ends of the exons together
80
Importance of Introns - Control
- Gene expression may be controlled by regulating
splicing - Alternate RNA splicing splicing at a different
location may code for a different protein by
removing different introns
81
- Exons code for domains in proteins
- Domains different functions on the same protein
- Rearrangement of exons may alter protein
shape/function - Crossing over during meiosis may create new gene
combinations - Variation evolution
82
Translation
- Decoding the mRNA and Converting the Information
Into Proteins
83
Anticodon
tRNA
Amino acid
84
Translation
- tRNA is interpreter mRNA sequence converted to
an amino acid sequence - Transfers amino acids from cytoplasmic pool to
the ribosome - Must recognize mRNA codons
85
(No Transcript)
86
- One end attaches to a SPECIFIC amino acid
- The other end contains a sequence of amino acids
that compliments the codon of the mRNA - Anticodon
87
- tRNA is manufactured in the nucleus by DNA code
- Can be used repeatedly
- Single-stranded
- Folded due to hydrogen-bonding
88
(No Transcript)
89
Translation tRNA
- 45 types of tRNA only 64 codons on the mRNA
- ? not enough types of tRNA for the mRNA codons
- 3rd base-pairing is loose
- tRNA can recognize 2 or 3 different mRNA codons
- WOBBLE
90
Wobble
- U in wobble position of tRNA can recognize A or G
- CCI (inosine) on tRNA can base-pair with U, C or
A on mRNA - GGU, GGC, GGA (mRNA) glycine
91
Wobble Genetic Variation Genetic Variation
Evolution
92
Redundancy more than one codon for amino acids
Redundancy prevents ?
Goofs
93
Translation
- Enzyme attaches specific amino acid
- Aminoacyl-tRNA synthetase
- Requires ATP
94
Ribosome
- 60 rRNA, 40 proteins
- Coordinates the pairing of mRNA and tRNA
- Binding site for mRNA and tRNA
- Two parts
- Small and large subunits
- Very specific ENZYMES
95
Large subunit
Binding sites
Small subunit
96
- mRNA and 3 tRNA binding sites
- P site polypeptide chain
- A site aminoacyl binding site
- E site exit
97
(No Transcript)
98
Translation
- Initiation
- Elongation
- Termination
99
Translation Initiation
- Small subunit binds to mRNA upstream from AUG
- AUG (mRNA) UAC (tRNA)
- Codes for Methionine START
- Large subunit attaches (GTP)
- P-site
100
(No Transcript)
101
(No Transcript)
102
Translation Elongation
- A-site available for next tRNA
- Codon recognized
- Peptide bond forms between amino acids (peptidyl
transferase) - Translocation tRNA shifts over to the P-site
freeing up the A-site for the next tRNA
103
Translation Termination
- Elongation continues (60 milliseconds per
transfer) - STOP codon
- UAA, UAG, UGA
- Release factor
- Enzyme releases polypeptide from P-site tRNA
104
(No Transcript)
105
Translation
106
Free and Bound (ER) Ribosomes
- Free polypeptides remain
- Bound secretion
- mRNA has a signal sequence that allows the
ribosome to bind to the ER - Polypeptide chain then grows into the cisternal
space of the ER
107
Polyribosomes
- mRNA can be read and translated by many ribosomes
simultaneously - Why?
108
(No Transcript)
109
From Polypeptide to Functional Protein
- Some polypeptides must be further modified to
become functional - Glycoproteins, lipoproteins
- Divided into smaller pieces insulin (quaternary
structure)
110
Prokaryotes vs. Eukaryotes
- Prokaryotes
- No separation of replication, transcription,
translation - Translation may begin before transcription is
finished - Eukaryotes have compartments so RNA can be
modified much more control
111
Prokaryote/Eukaryote protein synthesis
112
(No Transcript)
113
Mutations
- Permanent change in DNA
- Point mutation 1 or 2 nucleotides are incorrect
- 3 types
- Base-pair substitutions
- Insertions
- Deletions
- Frame-shift
114
Mutations Single Base-Pair
- Little or no damage
- Redundancy
- Wobble
- Can cause problems sickle-cell
115
Base-Pair Mutations
- Result in an improved protein (evolution) or
- Less active or inactive protein
- Missense altered codons make sense but not
intended - Nonsense can result in premature termination
nonfunctional protein
116
Insertion Mutations
- Insertion insertion of 1 or more base-pairs
- Deletion deletion of base-pair(s)
- Alters the triplet grouping (codon)
- Frameshift
117
(No Transcript)
118
Mutations Frame-shift
- THE DOG BIT THE CAT wild
- THE DOG BIT THE CAR substitution
- THE OG BIT THE CAT deletion
- THE OGB ITT HEC AT Frame shift
119
Mutagenesis
- Creation of mutations
- Spontaneous or
- Exposure to MUTAGENIC AGENTS
- Radiation, chemicals, viruses
- Carcinogenic
120
Evolution
121
Sequence of events in protein synthesis
122
(No Transcript)