classical genetics - PowerPoint PPT Presentation
1 / 25
Title: classical genetics
1
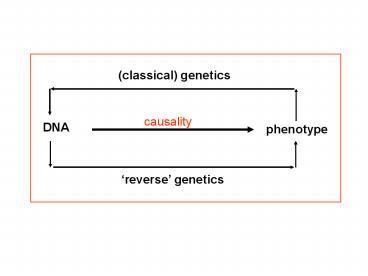
(classical) genetics
causality
DNA
phenotype
reverse genetics
2
Relationship between efficiency of mutation and
the number of clones required to guarantee
recovery of the required mutant, with 90
probability.
3
mismatch positions
Exchanging restriction fragments
4
use of inosine at randomized positions
5
Mutagenesis by mismatch primer extension on
single-stranded template.
T
C
6
Single or multiple point mutations, insertion
mutations and deletion mutations are possible.
Main concern is the efficiency of screening.
7
Theoretical concept, yielding 50 wild-type and
50 mutant progeny. but also single-stranded
templates transform the host, producing more
wild-type clones repair mechanisms favor
the original sequence.
8
Role of GATC sequences in mismatch repair
strand selectivity
9
Method of Eckstein
Introducing an S-modified strand. Some
restriction enzymes are inhibited by
S-modification, but still cleave the unmodified
strand (gt nicking) Exonuclease III removes the
mismatch from one strand only.
10
The structure of dCTPaS the Sp isomer is used
specifically by E. coli DNA polymerase I.
11
The biochemistry behind the method of Kunkel for
site-specific mutagenesis
Metabolism of dUTP in E. coli. Arrows indicate
directions of flow within the overall pathway
rather than the equilibrium of individual reaction
s. Bold arrows indicate the major pathway.
Boxes contain the structural genes of interest
ung uracil-DNA N-glycosylase dut
dUTPase dcd dCTP deaminase cdd
(deoxy)cytidine deaminase deoA thymidine
phosphorylase (deoxyuridine)
dCyd deoxycytidine dUrd deoxyuridine dThd
deoxythymidine dRib-1-P deoxyribose-1-phospha
te
12
Comparing the methods of Eckstein and Kunkel.
Kunkel gt the template strand is
modified gt "manipulations" in vivo Eckstein
gt the new strand is modified gt
manipulations in vitro
13
The gapped duplex method of in vitro mutagenesis
Conversion of host between amber suppressing
and non-suppressing phenotype. Selection by
using non-suppressing host.
14
Gapped duplex method with phenotypic selection
Switching between two resistance genes with
respect of suppressing phenotypes.
Not in 2009-2010
15
Selection of mutants by an extra mutation in
the bla gene to create ceftazidime resistance.
Not in 2009-2010
16
PCR-based methods for site-specific mutagenesis
Mismatch position in PCR primer cassette
approach Procedure adapted from SOE (splicing by
overlap extension) Megaprimer mutagenesis
Inverse PCR with point mutation(s), insertions
or deletions. gt stepwise synthesis of large
sequences feasible gt "universal" procedure
with classII-S restriction enzymes Mutant
selection with DpnI (the " ExciteTM "
method) Screening mutants via in vitro
methylation (the " GeneTailor TM " method) ...
many other alternatives, including multiple
site-directed mutations in coding sequences,
insertion unusual amino acids, gene shuffling,
etc.
Not in 2009-2010
Not in 2009-2010
Not in 2009-2010
17
18
The megaprimer method
The mutant molecules produced in the early rounds
of PCR act as primers for the later cycles of the
PCR.
19
Inverse PCR adaptations
Not in 2009-2010
20
Use of EarI or other ClassII-S enzymes
Not in 2009-2010
21
The ExciteTM method
Not in 2009-2010
22
The GeneTailorTM method
Not in 2009-2010
23
Making fusions between gene-1 and gene-2
Not in 2009-2010
24
25
(No Transcript)








![[Download ]⚡️PDF⚡️ The Cartoon Guide to Genetics (Updated Edition) PowerPoint PPT Presentation](https://s3.amazonaws.com/images.powershow.com/10072797.th0.jpg?_=20240704087)





















