TECHNIQUES IN MOLECULAR BIOLOGY - PowerPoint PPT Presentation
1 / 42
Title:
TECHNIQUES IN MOLECULAR BIOLOGY
Description:
TECHNIQUES IN MOLECULAR BIOLOGY CENTRIFUGATION- Separation of molecules/macromolecules/organelles according to the size, shape, density & gradient – PowerPoint PPT presentation
Number of Views:2037
Avg rating:3.0/5.0
Title: TECHNIQUES IN MOLECULAR BIOLOGY
1
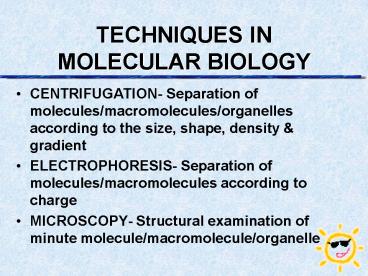
TECHNIQUES IN MOLECULAR BIOLOGY
- CENTRIFUGATION- Separation of molecules/macromolec
ules/organelles according to the size, shape,
density gradient - ELECTROPHORESIS- Separation of molecules/macromole
cules according to charge - MICROSCOPY- Structural examination of minute
molecule/macromolecule/organelle
2
CENTRIFUGATION
- MATERIALS OR PARTICLES IN A SOLUTION CAN BE
SEPARATED BY A CENTRIFUGE THAT USES THE PRINCIPLE
OF CENTRIFUGATION - CLASSES
- -ANALYTICAL/PREPARATIVE
- -ULTRACENTRIFUGATION AND LOW SPEED
- -DIFFERENTIAL/ZONAL CENTRIFUGATION
- http//ntri.tamuk.edu/centrifuge/centrifugation.ht
ml
3
ANALYTICAL CENTRIFUGATION
- IS USED TO MEASURE THE SEDIMENTED PARTICLE
PHYSICAL CHARACTERISTICS SUCH AS SEDIMENTATION
COEFFICIENT AND MOLECULAR WEIGHT
4
PREPARATIVE CENTRIFUGATION
- TO SEPARATE SPECIFIC PARTICLES THAT IS REUSABLE
- TYPES
- - RATE ZONAL
- - DIFFERENTIAL
- - ISOPYCNIC CENTRIFUGATION
5
ULTRACENTRIFUGATION AND LOW SPEED
- DEPENDS ON SPEED
- ULTRACENTRIFUGATION - THE SPEED EXCEEDS 20,000
RPM - SUPER SPEED ULTRACENTRIFUGATION- THE SPEED IS
BETWEEN 10,000 RPM-20,000 RPM - LOW SPEED CENTRIFUGATION- THE SPEED IS BELOW
10,000 RPM
6
DIFFERENTIAL CENTRIFUGATION
- PARTICLES IN SAMPLE WILL SEPARATE INTO
SUPERNATANT AND PELLET OR IN BOTH DEPENDING ON
THEIR SIZE, SHAPE, DENSITY AND CENTRIFUGATION
CONDITION - THE PELLET CONTAINS ALL THE SEDIMENTED COMPONENT
MIXTURE AND CAN CONTAIN MATERIALS THAT WAS NOT
SEDIMENTED EARLIER
7
DIFFERENTIAL CENTRIFUGATION
- SUPERNATANT CONTAINS MATERIALS THAT ARE NOT
SEDIMENTED BUT CAN BE SEDIMENTED WHEN
CENTRIFUGATION IS DONE AT A HIGHER SPEED
8
DIFFERENTIAL CENTRIFUGATION
9
ZONAL CENTRIFUGATION
- SAMPLE IS APPLIED ON TOP OF SUCROSE OR CESIUM
CLORIDE SOLUTION - PARTICLE CAN BE SEPARATED ACCORDING TO SIZE
SHAPE (TIME-RATE ZONE) OR DENSITY (ISOPYCNIC)
10
RATE-ZONAL CENTRIFUGATION
11
ISOPYCNIC-ZONAL CENTRIFUGATION
12
SEDIMENTATION COEFFICIENT
- WHEN CELL COMPONENTS ARE CENTRIFUFED THROUGH A
GRADIENT SOLUTION, THEY WILL SEPARATE INTO THEIR
OWN ZONE OR LINE/LAYER - THE RATE WHEN THE COMPONENT SEPARATES IS CALLED
AS SEDIMENTATION COEFFICIENT OR THE s VALUE
(SVEDBERG UNIT ) - 1 S 1 X 10-13 SECONDS
13
SEDIMENTATION COEFFICIENTVALUES
- PARTICLE OR SEDIMENTATION
- MOLECULE COEFFICIENT
- LYSOSOME 9400S
- TOBACCO MOSAIC VIRUS 198S
- RIBOSOME 80S
- RIBOSOMAL RNA MOLECULE 28S
- tRNA MOLECULE 4S
- HEMOGLOBIN MOLECULE 4.5S
14
SPEED OF CENTRIFUGATION
- A PARTICLE THAT IS ROTATING WILL HAVE A PULLING
FORCE IN A FORM OF MAGNITUDE TO SPEED FUNCTION AT
DEFINED ANGLE (ROTATION SPEED) AND CENTRFUGATION
RADIUS (THE DISTANCE BETWEEN THE SAMPLE CONTAINER
AND THE ROTOR CENTRE)
15
SPEED OF CENTRIFUGATION
- 2 WAYS OF EXPRESSING THE PULLING FORCE
- a) RELATIVE CENTRIFUGATIONAL FORCE-RCF (g)
- b) ROTATION PER MINUTE (rpm)
16
RELATIVE CENTRIFUGATIONAL FORCE
- THE PULLING FORCE OF CENTRIFUGATION IS BASED ON
OR RELATIVE TO THE STANDARD GRAVITATIONAL FORCE - FOR EXAMPLE 500x g MEANS THAT THE PULLING FORCE
IS 500 TIMES BIGGER THAN THE STANDARD
GRAVITATIONAL FORCE
17
RELATIVE CENTRIFUGATIONAL FORCE
- EQUATION
- R.C.F. 1.119 x 10 -5 (rpm2) r rpmrotation
per minute - rradius (in cm)
- UNIT g
18
ELECTROPHORESIS
- THE MOVEMENT OF CHARGED PARTICLE IS INFLUENCED BY
ELECTRICAL CURRENT - ELECTROPHORESIS IS THE METHOD OF SEPARATING
MACROMOLECULE SUCH AS NUCLEIC ACID AND PROTEIN
ACCORDING TO SIZE, ELECTRICAL CHARGE AND PHYSICAL
PROPERTIES SUCH AS DENSITY ETC - SEPARATION IS AIDED BY A MATRIX SUCH AS
POLIACRYLAMIDE OR AGAROSE
19
ELECTROPHORESIS
- PRINCIPLE SEPARATION OF MACROMOLECULE DEPENDING
ON TWO PROPERTIES WEIGHT AND CHARGE - ELECTRICAL CURRENT FROM THE ELECTRODE WILL PUSH
THE MOLECULE AND AT THE SAME TIME THE OTHER
ELECTRODE WILL PUT IT - MOLECULES WILL MOVE ALONG THE PORES THAT ARE
FORMED BETWEEN THE INTER-WOVEN MATRIX THAT ACTS
LIKE A SIEVE TO SEAPARATE THE MOLECULE ACCORDING
TO THEIR SIZE
20
ELECTROPHORESIS
- ELECTRICAL CURRENT WILL FORCE THE MACROMOLECULE
TO MOVE ALONG THE PORES - THE MACROMOLECULE MOVEMENT DEPENDS ON THE
ELECTRICAL FIELD FORCE, THE MOLECULE SIZE AND
SHAPE, THE SAMPLE RELATIVE HYDROPHOBIC PROPERTY,
IONIC STRENGTH AND THE TEMPERATURE OF THE
ELECTROPHORESIS BUFFER - DYEING WILL AID THE VISUALISATION OF
MACROMOLECULE IN THE FORM OF SEPARATED SERIES OF
STRIPES
21
PROTEIN ELECTROPHORESIS
- PROTEIN HAS A POSITIVE OR NEGATIVE NET CHARGE AS
A RESULT OF THE COMBINATION OF CHARGED AMINO
ACIDS CONTAINEDIN THEM - THE MATRIX THAT IS USUALLY USED FOR PROTEIN
SEPARATION IS POLIACRYLAMIDE - TWO DIMENSIONAL GEL ELECTROPHORESIS- PROTEIN
SEPARATION ACCORDING TO ISOELECTRICAL POINTS AND
MOLECULAR WEIGHT
22
2-D PROTEIN ELECTROPHORESIS
- FIRST STEP/DIMENSION
- PROTEIN SEPARATION ACCORDING TO ISOELECTRIC
POINT (PROTEIN CONTAINS DIFFERENT POSITIVE AND
NEGATIVE CHARGE RATIO) - -ELECTROPHORESIS IS DONE ON THE GEL IN THE FORM
OF TUBE PROTEIN WILL MOVE IN A SOLUTION WITH
DIFFERENT pH GRADIENT
23
2-D PROTEIN ELECTROPHORESIS
- FIRST STEP/DIMENSION
- -PROTEIN WILL STOP WHEN IT REACHES THE pH WHICH
IS EQUAL TO ITS ISOELECTRIC POINT i.e WHEN THE
PROTEIN DOES NOT HAVE A NET CHARGE.
24
2-D PROTEIN ELECTROPHORESIS
- SECOND STEP/DIMENSION
- PROTEIN SEPARATION BY MOLECULAR WEIGHT
- ELECTROPHORESIS IS DONE IN AN ORTHOGONAL
DIRECTION FROM - THE FIRST STEP
- SODIUM DODECYL SULPHATE
- (SDS) IS ADDED
25
2-D PROTEIN ELECTROPHORESIS
26
1-D PROTEIN ELECTROPHORESIS
- PROTEIN IS SEPARATED BY ITS MOLECULAR WEIGHT ONLY
- THE TECHNIQUE IS ALSO KNOWN AS POLIACRYLAMIDE GEL
ELECTROPHORESIS (PAGE) OR SDS-PAGE IF SDS IS
PRESENT DURING SAMPLE PREPARATION - SIMULATION OF 1-D ELECTROPHORESIS
- http//www.rit.edu/pac8612/electro/
- Electro_Sim.html
27
SDS-PAGE
- TO SEPARATE PROTEIN WITH THE SIZE OF 5 - 2,000
kDa - PORES IN BETWEEN THE POLIACRYLAMIDE MATRIX CAN
VARIES FROM 3-30 - THE PROTEIN SAMPLE IS IN THE FORM OF PRIMARY
STRUCTURE (SAMPLE IS BOILED WITH SDS AND
?-MERCAPTOETHANOL PRIOR BEING LOADED ONTO GEL)
28
SDS-PAGE
- PROTEIN IS STAINED USING COOMASIE BLUE OR SILVER
- NON-DIRECTIONAL STAINING CAN BE DONE
- -ANTIBODY BOUND WITH RADIOISOTOPE OR ENZYME,
FLUORESENCE DYE
29
SDS-PAGE
- SDS FUNCTION
- NEGATIVELY CHARGED DETERGENT THAT
- BINDS TO THE
- HYDROPHOBIC REGION
- OF THE PROTEIN
- MOLECULE AS A
- RESULT THE PROTEIN BECOMES A LONG POLIPEPTIDE
CHAIN AND FREE FROM OTHER PROTEINS AND LIPIDS
30
SDS-PAGE
- ?-MERCAPTOETHANOL FUNCTION TO BREAK DISULPHIDE
BONDS SO THAT PROTEIN SUBUNIT CAN BE ANALYSED
31
NUCLEIC ACID ELECTROPHORESIS
- AGAROSE OR POLIACRYLAMIDE IS THE MATRIX USUALLY
USED TO SEPARATE NUCLEIC ACID IN A TECHNIQUE
KNOWN AS AGAROSE GEL ELECTROPHORESIS - SAMPLE CONTAINING DNA IS LOADED INTO WELLS
LOCATED NEAR TO THE NEGATIVELY CHARGED ELECTRODE - DNA THAT IS NEGATIVELY CHARGED WILL BE ATTRACTED
TO THE POSITIVE ELECTRODE
32
NUCLEIC ACID ELECTROPHORESIS
- DNA WITH A BIGGER SIZE WILL MOVE SLOWER THAN THE
SMALLER SIZE WHICH MOVE FASTER - STAINING IS DONE USING ETHIDIUM BROMIDE (EtBr)
THAT ENABLES THE VISUALISATION OF NUCLEIC ACID
EtBr IS INSERTED BETWEEN THE BASES ON THE NUCLEIC
ACID - EtBr IS ORANGE IN COLOUR WHEN LIT-UP BY
ULTRA-VIOLET LIGHT
33
NUCLEIC ACID ELECTROPHORESIS
34
MICROSCOPY
- ONE OF THE EARLIEST TECHNIQUE TO STUDY
MACROMOLECULE - PRINCIPLE TO ENLARGE SMALL IMAGES
- TYPES OF MICROSCOPY ACCORDING TO THE SIZE OF
IMAGE ENLARGEMENT - - LIGHT MICROSCOPE (300nm-2mm)
- - ELECTRON MICROSCOPE
- (0.15nm-100?m)
35
LIGHT MICROSCOPE
- IMAGE ENLARGEMENT PRINCIPLE
- LIGHT FROM BELOW OF THE MICROCOPE GOES THROUGH
- THE CONDENSOR TO FOCUS THE
- LIGHT TO THE SPECIMEN.
- LIGHT FROM THE SPECIMEN IS
- RECOLLECTED BY THE OBJECTIVE LENSE TO FORM AN
IMAGE
36
LIGHT MICROSCOPE
- TYPES OF LIGHT MICROSCOPE
- BRIGHT-FIELD MICROSCOPE DARK-FIELD
MICROSCOPE - PHASE-CONTRAST MICROSCOPE
- FLUORESENCE MICROSCOPE (UV)
- (FLUORESCIN/RHODAMIN)
37
ELECTRON MICROSCOPE
- PRINCIPLE
- -ELECTRON IS USED (NOT LIGHT) TO ENLARGE IMAGE
- -SPECIMEN MUST UNDERGO A SERIES OF PREPARATION
PROCESSES SUCH AS COATING WITH THIN LAYER OF GOLD
TO ALLOW EMITTED ELECTRON TO COLLIDE TO AND THEN
RECOLLECTED TO FORM IMAGE ON THE SCREEN
38
ELECTRON MICROSCOPE
- TYPES
- 1) TRANSMISSION ELECTRON MICROSCOPE
- -ELECTRON GOES THROUGH THE SPECIMEN AND IMAGE IS
RECOLLECTED ON A FLUORECENS SCREEN - -THE INNER STRUCTURE OF THE SPECIMEN CAN BE SEEN
39
ELECTRON MICROSCOPE
- TYPES
- 2) SCANNING ELECTRON MICROSCOPE
- -ELECTRON IS FOCUSSED TO THE SPECIMEN AND THEN
REEMITTED (SCANNED) TO THE DETECTOR AND IMAGE IS
SEND TO THE SCREEN FOR VIEWING - -THE OUTER STRUCTURE CAN BE SEEN
40
ELECTRON MICROSCOPE
SCANNING ELECTRON MICROSCOPE
MOSQUITO IMAGES BY SCANNING ELECTRON MICROSCOPE
41
OTHER TECHNIQUES
- CHROMATOGRAPHY
- -PAPER PROTEIN SEPARATION BY USING FILTER
PAPER AS THE MATRIX - -ION-EXCHANGE
- -GEL FILTRATION
- -AFFINITY
- -HIGH PRESSURE LIQUID CHROMATOGRAPHY (HPLC)
42
OTHER TECHNIQUES
- RADIOISOTOPES FOR MOLECULE TAGGING 32P, 131I,
35S, 14C, 45Ca, 3H - - RIA, PULSE-CHASE EXPERIMENT,
AUTORADIOGRAPHY - ANTIBODY (MONOCLONE/POLYCLONE) FOR TAGGING
MOLECULE EIA, IF, ELISA - X-RAY DIFFRACTION ANALYSIS PROTEIN STRUCTURE
DETERMINATION - DNA RECOMBINANT TECNOLOGY