Bioinformatics: - PowerPoint PPT Presentation
1 / 15
Title:
Bioinformatics:
Description:
Title: PowerPoint Presentation Author: ACC Last modified by: Kim Minor Created Date: 4/25/2002 1:53:49 PM Document presentation format: On-screen Show – PowerPoint PPT presentation
Number of Views:123
Avg rating:3.0/5.0
Title: Bioinformatics:
1
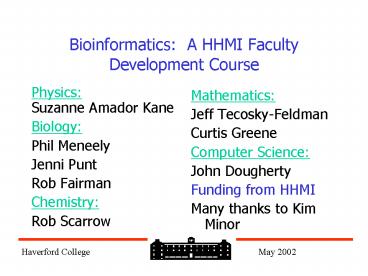
Bioinformatics A HHMI Faculty Development
Course
- Physics
- Suzanne Amador Kane
- Biology
- Phil Meneely
- Jenni Punt
- Rob Fairman
- Chemistry
- Rob Scarrow
- Mathematics
- Jeff Tecosky-Feldman
- Curtis Greene
- Computer Science
- John Dougherty
- Funding from HHMI
- Many thanks to Kim Minor
Haverford College May 2002
2
Course Goals
- Its unrealistic for us to hire a bioinformatics
faculty member or teach entirely new courses on
the subject, so we instead chose to - Educate faculty about theory behind and use of
bioinformatics tools - Stimulate development of research applications
- Stimulate use of bioinformatics in courses
- Get faculty talking across departmental
boundaries
Haverford College May 2002
3
Course Format
- Eleven weekly 1 ½ lectures
- Six 1 ½ hour hands-on computer workshops
- Dinner with instructors in between lecture
workshop - Informal homework graded by instructors
- Assigned readings lecture notes
- Follow-up subgroup meetings throughout the spring
term - Faculty received one course stipend or release
Haverford College May 2002
4
The Instructors
- Greg Grant
- Elisabetta Manduchi
- Warren Ewens
- U. Penn. Center for Bioinformatics
- Guest lecturers
- Jessica Carol Kissinger
- Jonathan Crabtree (both of U.Penn.)
- Stephen Bryant (NIH National Center for
Biotechnology Information)
Haverford College May 2002
5
Course Content Syllabus
- Topics covered
- Statistics
- Sequence alignment
- Gene finding
- Gene expression/DNA microarrays
- Phylogenetic analysis
- Protein structural alignment
- Kim Minor has made a very comprehensive course
website - http//www.haverford.edu/biology/genomicscourse/ge
nomics.htm
Haverford College May 2002
6
Textbooks, software, other resources
- We have compiled a list of useful textbooks,
Powerpoint presentations, and course materials
from other institutions. - Lists of and links to these resources are
available on the course website.
Haverford College May 2002
7
Research applications
- Phil Meneely, Curtis Greene Rob Manning will
perform a phylogenetic analysis of the RCC-1
protein family this summer with rising junior
Ethan Roland. - John Doughertyongoing research with David Barkan
(HC undergrad) on A Parallel Implementation of
the Needleman-Wunsch Algorithm for Global Gapped
Pair-Wise Alignment. (poster presented April
2002 at Consortium for Computing at Small
Colleges 7th Annual NE Conference.) - Jenni Punt will perform a PSI-BLAST analysis of
CDK2 in order to understand possible targets for
its unexpected role in programmed cell death
Haverford College May 2002
8
Current curricular development
- Bio 100 What is life? Phil Meneely BLAST and
clustalw used to understand phylogenetic
relationships of beta globins of humans and other
mammals. - Bio 200 Cellular Molecular Biology. Jenni
Punt DNA Microarray/Gene expression
examplestumor classification Rob Fairman
Sample BLAST analysis of a-amylase. - Physics 320 Intro Biophysics Suzanne Amador
Kane sequence analysis, DNA chip technology for
gene expression, protein structual prediction) - CS392 Parallel Computing J.D. assignment
implementing a sequential and parallel version
of the Needleman-Wunsch algorithm for DNA
sequence alignment. - Math 218 Probability Statistics Curtis
Greene dishonest casino example of hidden
Markov chains probability statistics of
sequence alignments.
Haverford College May 2002
9
Ongoing curricular development
- Bio 303 Protein Structure/Function Rob Fairman
New modules on PSI-BLAST sequence analysis and
protein structural alignments (threading
algorithms) - Bio 300 PSI-BLAST lab module on anthrax porins
- CS206 Data StructuresJ.D. Assignment on
phylogeny to provide experience with trees and
matrices in a bioinformatics context - Chem 351 Bioinorganic Chemistry Rob Scarrow
J.D. phylogenetic analysis of metalloproteins
to understand how the binding of different metals
has evolved. - Bioinformatics will be an important part of a new
biochemistry biophysics Superlab course
Haverford College May 2002
10
Conclusions
- The Faculty Development course structure has been
very successfully applied in Computing across
the Sciences (Spring 2001) and Bioinformatics
(Fall, 2001). - Faculty participation has been strong and
consistent. - New research and curricular applications have
resulted as a consequence. - The next installment will be a course next spring
on Science and Society which we hope will draw
in faculty from other divisions.
Haverford College May 2002
11
- Faculty enjoyed the chance to meet and talk
across disciplinary divisions - I felt like learning about different teaching
styles and different approaches to science was
the most important thing to come out of this
experience.
Haverford College May 2002
12
- Positive outcomes include my understanding of
bioinformatics, and my appreciation for the use
of probability and statistics in biology. I also
consider the professional relationships forged in
the lab trying to"BLAST" protein sequences or
construct SQL queries a positive outcome. - I knew absolutely NOTHING about bioinformatics
prior to the course. I've learned a lot about the
tools and strategies, and the central problems of
a whole area of inquiry of which I was ignorant.
This was a terrific way to get to know colleagues
in the Biology dept. Our collaboration has
spawned other discussions about ways that the
Mathematics dept can serve the Biology major.
Haverford College May 2002
13
- Mainly, working cohesively with my colleagues
on bringing bioinformatics into our curriculum.
It has gotten me to appreciate how we can work
together as a Department on such goals and to
articulate the need for expertise in this area
for the long-term goals of this Department. - I wanted to find out what all the fuss was
about. I had some general ideas about what
bioinformatics could do and what tools were out
there, but I'd only used one or two and I wasn't
sure how they worked. I realized that the things
that will be most useful to me are things I'd
already heard of, but I was glad for the
increased level of under-standing of the
bioinoformatic tools. It was great to get
together with other faculty in working toward a
common intellectual project.
Haverford College May 2002
14
- On a more cosmic level, I am now much more
"available" to offer advice to seniors who are
thinking about research problems in this area. I
can imagine it leading to specific mathematical
projects in the future (for example, studying the
statistics in Greg and Warren's book). - I understand the power of PSI-BLAST a lot more
now. I would never have thought of this research
project without the course.
Haverford College May 2002
15
- Drawbacks
- Time constraints in working out the examples and
doing background reading - Time constraints in implementing curricular
development. - Requests for more hands-on workshops.
- More application-driven emphasis to math stats
(less abstraction).
Haverford College May 2002