Bacteriology Lecture 7 PowerPoint PPT Presentation
1 / 39
Title: Bacteriology Lecture 7
1
Transcription the players
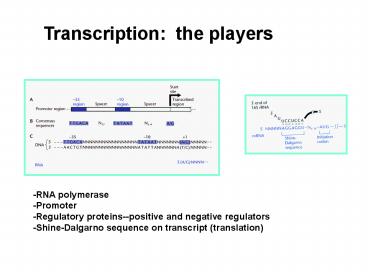
-RNA polymerase -Promoter -Regulatory
proteins--positive and negative
regulators -Shine-Dalgarno sequence on transcript
(translation)
2
Transcription is the primary site of control in
prokaryotes Prokaryotic genes are arranged in
operons Promoters drive the expression of genes
3
Coordinate Regulation
- Expression of several or numerous genes can be
controlled simultaneously. - Operon a set of genes that are transcribed
from the same promoter and controlled by the same
regulatory sites. - Regulon a set of genes (and/or operons)
expressed from separate promoter sites, but
controlled by the same regulatory molecule.
Global regulons may coordinate expression of
many genes and operons, and may induce some, but
repress others.
4
Other sigma factors
5
Alternate Sigma Factors
recognize promoters of different architecture
different regulons of genes
Ishihama, 2000, Ann. Rev. Microbiol. 54499-518
6
Sigma factors control of regulons
- Varied by synthesis of new ??subunits that cause
RNA polymerase to recognize different promoter
sequences. - 1. The heat shock regulon responds to a rise in
temperature or chemical stress. - In E. coli, synthesis of ? 32 (the normal ? is a
?70) results in increased expression of at least
20 gene products. Many are chaperone proteins or
proteases which counteract the affects of high
temperature on protein folding and
protein-protein interactions. - 2. Synthesis of 9 new ? factors are involved in
the sequential triggering of events required for
sporulation of Bacillus spp.
7
(No Transcript)
8
Enhancers
9
A s54 dependent activation mechanism (the
odd-ball sigma) Mediated through interaction
with the NtrC transcription activator an
enhancer-type protein NtrC stimulates s54 RPO
formation therefore affects kII
Rippe et al. 1997, J. Mol. Biol. 270135-138
10
Most s factors are in the s70 family and hence
function like s70 s54 is different must be
activated by activator proteins after binding
kII is affected RPO is formed
Gralla, 2000, J. Bacteriol. 1824129-4136
11
Transcription factors
Typically DNA binding proteins that associate
with the regulated promoter and either decrease
or increase the efficiency of transcription,
repressors and activators, respectively - A
significant number of regulators do either one
depending on conditions
Brock Biology of Microorganisms, vol. 9, Chapter 7
12
Gene expression responds to environment
Arginine biosynthesis
Lactose degradation
Induction
Repression
Brock Biology of Microorganisms, vol. 9, Chapter 7
13
Repression
14
A repression or co-repression mechanism
Example arginine, one of 20 essential amino
acids bacteria can make their own, but if
supplied with arginine will not express the
biosynthetic gene (Why bother making it if their
is plenty around?)
No arginine Cell needs it Repressor does not
bind Arginine genes expressed
High arginine Cell does not need it Repressor
Arg bind Arginine genes not expressed
Arginine
Brock Biology of Microorganisms, vol. 9, Chapter 7
15
Negative control of tryp enzymes
- If tryptophan is available, the allosteric
repressor protein (TrpR) binds to the operator
site, and blocks the promoter site. When
tryptophan is not available, the repressor cannot
bind, and RNA polymerase will initiate
transcription of the operon
16
Induction
17
An inducible repressor mechanism
Example lactose, sugar carbon (food)
source bacteria will utilize lactose if it is
present, but do not express genes if no lactose
is around (Why bother to make enzymes for using
lactose, if it is not present?)
No lactose Cell cannot use it Repressor binds to
lac genes No Lac enzymes are produced
Lactose present Cell can use it as
nutrient Repressor lactose fall off DNA Lac
enzymes are made
Lactose
Brock Biology of Microorganisms, vol. 9, Chapter 7
18
Gene expression responds to environment
Arginine biosynthesis
Lactose degradation
Induction
Repression
Brock Biology of Microorganisms, vol. 9, Chapter 7
19
(No Transcript)
20
(No Transcript)
21
Dual control Catabolite Repression
22
Transcriptional activation can occur via several
different mechanisms Almost always involves
contacts with RNAP
An excellent example regulates catabolite
repression catabolite activator protein
(CAP) A global regulator controlling gt 100
promoters
Brock Biology of Microorganisms, vol. 9, Chapter 7
23
(No Transcript)
24
(No Transcript)
25
Class I
CAP is activated by binding of cAMP
(intracellular levels elevated when low
glucose) Protein dimerizes and binds to
promoters in a number of different ways induces
a high angle bend in DNA Class I upstream of
-35 Class II overlaps with 35 Contacts with
RNAP stimulate transcription
Class II
Busby and Ebright, 2000, J. Mol. Biol. 293199-213
26
Models for Class I and Class II promoter
activation
Class I CAP binding sites can be from 62 to
103. CAP interact with the carboxy terminus of
the RNAP a-subunit (aCTD)
Class II CAP binding sites usually overlap the
35. CAP interact with the aCTD, aNTD, and the s
factor
Busby and Ebright, 2000, J. Mol. Biol. 293199-213
27
Activation
28
Transcriptional activation mechanism
Example maltose, sugar carbon (food)
source bacteria will utilize maltose if it is
present, but do not express genes if no maltose
is around (Why bother to make enzymes for using
lactose, if it is not present? same logic as
for lac)
No maltose Cell cannot use it Activator does not
binds DNA No Mal enzymes are produced
Maltose present Cell can use it as
nutrient Activator Maltose cab bind DNA Mal
enzymes are made
Promoter is weak
Maltose
Brock Biology of Microorganisms, vol. 9, Chapter 7
29
Attenuation
30
Attenuation
- Affects the transcription by adjusting the rate
at which transcripts are completed after they are
initiated. - When particular mRNA structures form,
transcription is likely to terminate. The rate
at which ribosomes progress behind the polymerase
will alter the RNA structures that can form.
Thus, the transcription of enzymes for synthesis
of an amino acid can be adjusted to the rate at
which the codons for the amino acid are
translated. - The trp operon regulated by a Repressor and by
Translational attenuation - Genes trpEDCBA encode enzymes for synthesis of
tryptophan
31
(No Transcript)
32
(No Transcript)
33
(No Transcript)
34
(No Transcript)
35
(No Transcript)
36
l Repressor prevents PR activity by steric
hindrance
Lytic Functions
-10
-35
-35
-10
Lysogenic Functions
OR2
OR1
OR3
PR
PRM
37
Attenuation
- The need for tryptophan is tested during
translation of the leader sequence (trpL) on
all mRNA initiated at P. - trp expression is modulated by the tRNA-trp
(charged tRNA) available to translate codon
UGG. If the ribosomes translate these codons
rapidly, transcription will stop because - Different segments in the leader mRNA sequence
can pair with each other to form alternative
stem-loop structures. - If ribosomes stall on the trp codons, the RNA
structure formed is not a rho independent
terminator. TRANSCRIPTION OF trp OPERON PROCEEDS - If ribosomes translate and pass trp codons
quickly, a terminator is formed. TRANSCRIPTION OF
trp OPERON TERMINATES
38
Signal Transduction
- Proteins that sense the environment, generate a
signal, transmit the signal through
protein-protein interaction and activate the
appropriate response regulon. - Bordetella pertussis
- Vibrio cholerae
39
Bacterial two-component systems
Bacterial two-component systems The sensor is
the transmembrane protein. Recognition of an
environmental signal activates a kinase
activity. This phosphorylates a histidine
residue on the sensor. The phosphate is
transferred to an aspartic acid residue on
another protein (the response regulator). The
phosphorylated regulator activates transcription
of the appropriate set of genes.