Presentacin de PowerPoint PowerPoint PPT Presentation
1 / 25
Title: Presentacin de PowerPoint
1
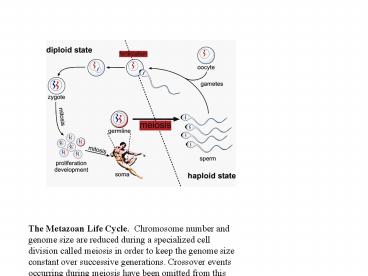
The Metazoan Life Cycle. Chromosome number and
genome size are reduced during a specialized cell
division called meiosis in order to keep the
genome size constant over successive generations.
Crossover events occurring during meiosis have
been omitted from this figure. Petronczki, et
al., 2003. Cell 112 423-440.
2
DNA STRUCTURE
- chemical structure of the 4 nucleotides in DNA
3
DNA STRUCTURE
- hydrogen bonded nucleotides on opposite helices
- DNA helices are antiparallel
- carbons on sugar define ends... 5' and 3'
- pyrimidines bond with purines
- T ? A
- C ? G
4
DNA STRUCTURE
- bonding is specific pyrimidines bond with
purines - implication means for accurate replication
5
DNA STRUCTURE
- helical structure of DNA
- major minor groves
- 10Å radius 20Å diameter
- 3.4Å between nucleotide base pairs
- 34Å / 360 turn
- 10 nucleotide base pairs / 360 turn
6
DNA REPLICATION
- 3 possible models of DNA replication
7
DNA REPLICATION
- model of replication proposed by Watson Crick
(1953) - parental strand template
- semiconservative model (new double helix has 1
template 1 new daughter strand) - replication fork
8
DNA REPLICATION
- replication fork ?
- bacteria incubated with 3H thymidine...
- daughter molecules ? 1 hot 1 cold strand
- autoradiograph...
9
DNA REPLICATION
- replication... theta (?) structures
- replication fork
10
MECHANISM OF DNA REPLICATION
- summary of DNA replication
- know structure, directionality, enzymes,
functions
11
MECHANISM OF DNA REPLICATION
- DNA replication requirements...
- 1. H bonds between bases must be broken
- 2. chain separation/unwinding
- 3. available pools of 4 dNTPs A T, C ? G
- 4. enzymes
- The minimal activities required at a replication
fork are the following a DNA helicase must
unwind the parental template a primase must
synthesize short oligoribonucleotides that serve
as primer for synthesis of the Okazaki fragments
on the lagging strand, a single-stranded
DNA-binding protein that coats the
single-stranded lagging-strand template and
interacts with other replication proteins and a
DNA polymerase must synthesize the nascent
leading and lagging strands. - E. coli...
- 4.7 107 bp (47,000,000)
- molecule has 4.7 106 turns (10 bp/turn)
- 1500 bp/s, 20 min to divide with 1 origin
12
MECHANISM OF DNA REPLICATION
- DNA polymerase I or pol I... several types of
activity - 1. polymerase catalyzes chain growth 5' ? 3'
- 2. exonuclease removes mismatches 3' ? 5'
- 3. exonuclease degrades double stranded DNA 3'
? 5'
13
MECHANISM OF DNA REPLICATION
- DNA polymerase III or pol III
- ... holoenzyme of 20 subunits
- 1. polymerase catalyzes chain growth 5' ? 3'
14
MECHANISM OF DNA REPLICATION
- prokaryotes have a single origin of replication
- bidirectional replication with 2 forks ? single
terminus - 20 minutes (... book says 40)
1000 nt/s (Chandler et al. 1975) less that one
mistake in 109 nucleotide polymerization events
(Drake et al. 1969).
Kornberg, A. and Baker, T.A. (1992). DNA
Replication. (New York W.H. Freeman)
15
MECHANISM OF DNA REPLICATION
- E. coli origin of replication
- tandem repeat consensus sequence
- protein binding sites
16
MECHANISM OF DNA REPLICATION
- replication to completion...
17
MECHANISM OF DNA REPLICATION
- eukaryotes have multiple origins of replication
- bidirectional replication with 2 forks
- 1.4 h (yeast) to 24 h (cultured animal cells)...
or more - issues...
- linear vs circular
- coordination of multiple chromosomes
- pulse/chase and autoradiography ...
18
MECHANISM OF DNA REPLICATION
- priming DNA synthesis
- leading lagging strands
- Enzymes
- Helicasa rompe los puentes de hidrogeno
19
MECHANISM OF DNA REPLICATION
- DNA structure
- replication
- enzymes
- Functions
- Topoisomerasa desenrolla el DNA
Fig 8-20
20
MECHANISM OF DNA REPLICATION
- DNA polymerase catalyzes replication 5' ? 3'
- extends but cannot initiate replication ...
Fig 8-20
21
MECHANISM OF DNA REPLICATION
Fig 8-27
- priming synthesis
- primase enzyme
- synth. short (30 bp) RNA primer
- primase DNA proteins primosome
22
MECHANISM OF DNA REPLICATION
Fig 8-28
- leading lagging strand
- replication is 5' ? 3'
- replication moves along template 3' ? 5'
- DNA pol III most synthesis
- DNA pol I fills gaps on lagging strand
- DNA ligase seals gaps
23
MECHANISM OF DNA REPLICATION
- replication fork
- lagging strand
- a) RNA primer growth
- b) DNA elongation as Okazaki fragments
- c) RNA primer removed by DNA pol I
- d) DNA ligation
24
Asymmetric Polymerase Action Is Determined by the
Helicase. The two polymerase cores within the
holoenzyme are shaded differently to show that
whichever orientation the holoenzyme assumes, the
resulting replisome is the same. Contact with the
DnaB helicase holds the leading strand core to
DNA, the lagging stand core remains free to
cycle.
Yuzhakov, et al., 1996. Cell 86 877-886.
25
One Possible Arrangement of Proteins at the
Replication Fork. In this diagram, the leading-
and lagging-strand polymerases are shown in an
anti-parallel orientation. With this symmetry,
only the t subunit associated with the
lagging-strand polymerase would likely be close
enough to DnaB to establish a proteinprotein
connection. See text for details. The drawing is
not to scale.
Kim, et al., 1996. Cell 84 643-650.