Carl Branden PowerPoint PPT Presentation
Title: Carl Branden
1
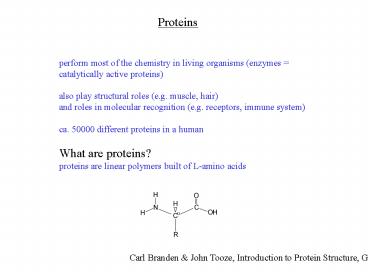
Proteins
perform most of the chemistry in living organisms
(enzymes catalytically active proteins) also
play structural roles (e.g. muscle, hair) and
roles in molecular recognition (e.g. receptors,
immune system) ca. 50000 different proteins in a
human What are proteins? proteins are linear
polymers built of L-amino acids
Carl Branden John Tooze, Introduction to
Protein Structure, Garland, 1998
2
Proteins are built of L-amino acids. 20 different
amino acids are encoded by specific DNA base
triplets. The amino acids are linked together by
amide bonds. Proteins are linear chains of amino
acids. Peptides are short proteins (lt 50
residues) Peptide bond is another name for the
amide bond connecting two amino acids.
dipeptide
Peptide bonds are planar due to partial
double- bond character. The backbone of a
protein consists of the atoms N, Ca and C. Side
chain carbons are labelled b, g, d, etc. Bond
lengths and bond angles are invariant. Only
dihedral angles vary. By convention, a protein
starts at the N-terminus and ends at the
C-terminus. (The N-terminus is synthesized first
during translation.)
3
glycine, Gly, G
alanine, Ala, A
hydrophobic A I L M F P W V positively charged
R K (H) negatively charged D E polar C N Q S T
Y tiny G
arginine, Arg, R
aspartic acid, Asp, D
asparagine, Asn, N
cysteine, Cys, C
glutamic acid, Glu, E
glutamine, Gln, Q
isoleucine, Ile, I
leucine, Leu, L
histidine, His, H
lysine, Lys, K
methionine, Met, M
phenylalanine, Phe, F
proline, Pro, P
serine, Ser, S
threonine, Thr, T
tryptophan, Trp, W
tyrosine, Tyr, Y
valine, Val, V
4
Hydrophobic cores are tightly packed
The interior of protein structures is tightly
packed. Water is excluded, except for very few
buried hydration water molecules. Almost all
residues in the interior are hydrophobic or, at
least, uncharged. Charged residues are on the
protein surface. The same general rules apply to
protein-protein binding surfaces. Regular
secondary structures form, because amide groups
are polar, seeking H-bonding partners when buried
in a hydrophobic environment.
CPK model of ubiquitin yellow hydrophobic grey
polar but uncharged blue positively charged red
negatively charged green backbone atoms
Backbone H-bonds
Only backbone and hydrophobic sidechains
retained
5
Protein structures are complicated. Simplified
representations emphasize the backbone.
myoglobin (stores O2 in the muscle)
6
a-helix 3.6 residues per turn, H-bonds between
residues i and i4
7
antiparallel b-sheet
parallel b-sheet
in both types of b-sheets, the side-chains point
alternatingly above and below the plane of the
sheet
8
mixed b-sheet
example thioredoxin
9
Primary, secondary, tertiary, quarternary
structure
Primary structure amino acid sequence Secondary
structure helices, sheets, turns (i.e. regular
sub-structures defined by H-bonds between
backbone amides) Tertiary structure 3D
structure Quarternary structure complex between
different protein molecules (e.g. dimer, trimer,
tetramer)
2 Cys residues can form a disulfide
bridge -CH2-SH 1/2 O2
-CH2-S-S-CH2- H2O
10
Prominent examples of drug-protein complexes
HIV protease inhibitors
COX2 inhibitors
cyclooxygenase activity
HIV must use HIV protease in order to cleave a
large polyprotein into functional proteins
essential to the structure of HIV and to its RNA
packaging.
PGG2
arachidonic acid
11
Rational drug design
drug molecules must fit snuggly in the enzymes
active site
influenza neuraminidase-inhibitor complex
12
Large ribosomal subunit from Haloarcula
marismortui
Science 289, 905 (2000)
13
Since peptide bonds are planar (and virtually
always trans), the backbone conformation of
each amino acid is determined by only two
dihedral angles, f and y. Knowledge of the f/y
pairs of each residue is sufficient to define
the 3D structure of the entire backbone! Due to
steric hindrance, the number of possible f/y
combinations is restricted.
14
Leventhals paradox
Assume a small protein with 100 amino acids, each
one of them can access 3 different
conformations 3100 5 x 1047 conformations Fastes
t motions 10-15 sec, so sampling
all conformations would take 5 x 1032 sec 60 x 60
x 24 x 365 31536000 3.1536 x 107 seconds in a
year Sampling all conformations will take 1.6 x
1025 years, much longer than the age of the
universe
In nature, proteins fold correctly within seconds!
The 3D structure is unambiguously encoded in the
amino-acid sequence, but protein structures are
very hard to predict from amino acid sequence,
unless the structure of a similar protein (gt 20
amino-acid sequence identity) is known.
Different proteins fold by different,
unpredictable mechanisms (some of them even need
helper proteins (chaperones) to fold. The
current picture is that of a folding funnel,
where the vertical axis displays energy and the
width of the funnel represents the accessible
conformational space.
15
From Structure to Function
Convergent evolution the overall structures of
chymotrypsin and subtilisin are very different,
but the catalytic triade (Asp, His, Ser
side-chains shown in blue) is conserved
subtilisin
chymotrypsin
16
Enzyme mechanism
how chymotrypsin digests proteins (peptide
hydrolysis)