mRNA Processing PowerPoint PPT Presentation
1 / 22
Title: mRNA Processing
1
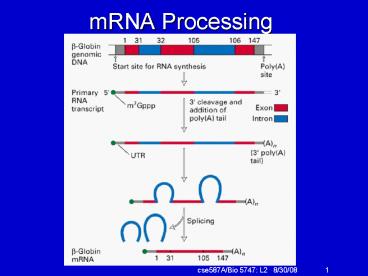
mRNA Processing
2
RNA splicing
- Splice sites are encoded in the sequence.
- Splice site recognition is complex and imperfect.
3
Alternative splicing
- Genes can produce multiple mRNA isoforms
- Human avg 5 mRNA/gene highly skewed
- Fewer introns/gene correlates w/ fewer
isoforms/genes
- UCSC genome browser http//genome.ucsc.edu
4
Genes
- Molecular definition
- DNA regions that are transcribed into a single
RNA strand, with nearby DNA regions controlling
time and quantity of transcription - Protein-coding genes and ncRNA genes
- Classical definition
- Whatever it is that gives rise to a heritable
trait
5
Genome sizes
- Widely varied
- Not well correlated with organism
complexity/sophistication - Typical bacterium 1-10 megabases (mb)
- Typical single-celled eukaryote 10-30 mb
- Smallest plants and animals 100 mb (fruit fly,
worm, mustard weed) - Human 3 gb some rats gophers 5-6 gb
- Pine tree 60 g Fern is 160 gb
- Database of Genome Sizes (DOGS)
6
Protein coding gene counts
- Not so widely varied
- E. coli (bacterium)
- 4,300 1200 nt/gene (no introns)
- S. cerevisiae (yeast)
- 5,800 2100 nt/gene (almost no introns)
- C. elegans (worm)
- 20,000 5000 nt/gene (5 introns/gene)
- Human other mammals
- 21,000 143,00 nt/gene (10 introns/gene)
7
Common human repeats
- LINES Long interspersed elements
- Most common is LINE1
- 6-7 kb
- 60,000 copies in human. 15 of genome.
- SINES Short interspersed elements
- Most common is Alu
- About 300 bp
- 1 million copies. 10 of human genome.
- Low complexity
- E.g., 5-10bp tandemly repeated repeated
- Especially near centromeres telomeres.
8
Computational problems Predicting exon-intron
structures
- De novo gene (mRNA) prediction Given only a
genome sequence, predict the exon-intron
structures of all mRNAs it encodes - Multi-genome de novo Given also the genome
sequences of 1 or more related organisms - RNA-based given also partial sequences of a
subset of RNAs from a variety of species - Difficult in plants animals because most
genomic DNA does not encode proteins
9
Computational Problems Categorizing proteins by
function
- After determining the mRNA products
- Relatively easy to infer the sequences of the
proteins they encode - Need to infer their approximate functions
- Search database of proteins with known function
Search Alignment Problem - Find domains (modular components) with known
function. E.g., - DNA binding, ATPase, transmembrane,
10
Non-coding RNA
- Functions
- Transfer RNAs codon-to-amino-acid adapters
- Ribosomes catalyze amino acid linkage
- Protein-RNA complex. RNA is catalytic!
- Small RNAs edit specific mRNAs, or
- prevent translation of specific mRNAs
- All transcribed from DNA but not translated
- Structure
- Shape, determined by self-pairing, is essential
- External base-pairing is usually essential, too
11
RNA
- Normally single-stranded
- Much less stable than DNA. Shorter lifetime.
- Can form complex structure by self-base-pairing
12
RNA self-base-pairing
13
3D shape of transfer RNA
14
Computational problems nc RNA
- secondary structure Given an RNA sequence,
predict its folded form -- which bases will pair
with one another - RNA homology Given an RNA sequence, search db
for sequences with the same secondary structure - Given an RNA sequence and its 2nd-ary structure,
search db for sequences with the same 2nd-ary
structure
15
Transcriptional regulation
- Transcription factors (TFs)
- Proteins that bind to short (8-16 nt) DNA
sequences, affecting transcription rate - In smaller genomes, TF binding sites are
typically in the promoter region, upstream of a
genes transcription start site - Repressors reduce the genes transcription rate,
activators increase it - Transcription is initiated by RNA Polymerase II
binding to the transcription start site
16
TF binding sites
- Specificity
- Each type of TF (300 in yeast) binds to a class
of sequences called a motif - Computational problems involving TF binding sites
- Find functional binding sites in genomic DNA
sequence for a TF with a known motif - Find functional binding sites without a known
motif - Given sequences containing sites for a given TF
find the sites and determine the motif
17
TFs form regulatory networks
- Glucose sensing signaling in yeast
18
Developmental gene regulation
Johnson et al. 2007, Science
19
Computational problems in systems biology
- Network identification
- Given, e.g., gene expression levels in a variety
of conditions, or a time series after stimulus,
infer the controlling TF network - There are various other data sources
- It helps if you know the TF binding motifs
- Network simulation
- Given a particular gene regulation network,
predict how it will respond to novel stimuli or
genetic manipulation, as a function of time
20
DNA Packaging
- DNA is packed hierarchically
- The chromosome is the largest package
- Humans have 22 chrs 2 sex chrs
- Human genome 2m long 0.34nm/base
- DNA is 1 picogram (10-12g) per gigabase
Quicktime animation
21
Epigenomics
- Chromatin marks
- Chemical modification of histones
- Affects DNA accessibility
- Specific marks correlated with transcription
initiation, elongation, and silencing - Code is not well understood
Peterson et al 2004
22
Epigenomics
- DNA methylation
- Mostly mCG, mCNG (plants)
- Heritable through cell division
- Associated with gene silencing
- Controls mobile elements
- X-inactivation and imprinting
- mC often mutates to T, leaving few CG pairs
- Except in rarely methylated regions (e.g.
promoters of genes essential to all cell types)