ChIPchip PowerPoint PPT Presentation
1 / 36
Title: ChIPchip
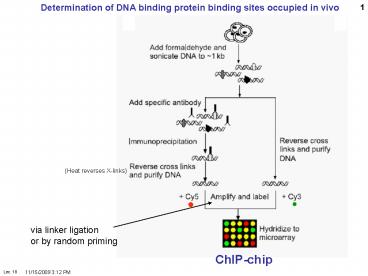
1
Determination of DNA binding protein binding
sites occupied in vivo
(Heat reverses X-links)
via linker ligation or by random priming
ChIP-chip
2
Cawley et al. Cell, 116, 499509, 2004,
3
(No Transcript)
4
High throughput gene expression
SAGE Serial Analysis of Gene Expression Provides
sequence information
A
B
A
B
5
B
A
(A type IIs restriction enzyme)
(e.g. NlaIII)
6
A type IIs restriction enzyme cuts outside its
recognition sequences
Fok I Flavobacterium okeanokoites
9
13
FokI cleavage site is to the right of the last N
in each strand above, generating a staggered cut
with a four base 5 overhang.
Ends are in general not compatible with other
FokI cut fragments, but are specific for a given
locus.
7
B
A
B
A
8
What Exactly is DNA Expression? DNA expression
refers to the study of how specific genes are
transcribed at a given point in time in a given
cell. A gene is transcribed into a messenger RNA
(mRNA) transcript when the protein that is
encoded by the gene is required by the cell. This
occurs because DNA located in the nucleus, but
all of the machinery necessary for translation,
or producing proteins, resides in the cytoplasm.
The cell resolves this problem by creating a copy
of the gene (mRNA) that is capable of entering
the cytoplasm through the nuclear pores. By
examining which transcripts are present in a
cell, it is possible to deduce which genes (and
their related proteins) are expressed in a cell
type, and at what time these are expressed. In
the past, DNA expression studies typically looked
at only a few transcripts at any one time, due to
the limitations of the techniques available1. But
in recent years several new techniques have been
developed that enable large scale studies of DNA
expression these can be used to create
'expression profiles'. An expression profile is a
characterization of the relative quantity of
every transcript that is produced in any one cell
type. One technique that has been used to
generate expression profiles is SAGE (Serial
Analysis of Gene Expression). What is SAGE
(Apart from a Spice) SAGE is a technique that
allows rapid, detailed analysis of thousands of
transcripts in a cell. The basic concept of SAGE
rests on two principles firstly, a small
sequence of nucleotides from the transcript,
called a 'tag', can effectively identify the
original transcript from whence it came, and
secondly, that linking these tags allows for
rapid sequencing analysis of multiple
transcripts. Imagine having thousands of
transcripts to sequence - each sequencing event
would take a certain amount of time to complete,
and several thousand of these events would be
necessary to identify each individual transcript.
By linking the tags together, only one sequencing
event is required to sequence every transcript
within the cell, making the task of DNA
expression profiling a much less daunting
one1,2. Five Easy Steps and You Too Can Do
SAGE Figure 2 shows a schematic diagram of each
of the steps in SAGE. First, a complimentary DNA
strand, or cDNA, of each transcript in the cell
must be generated. This is necessary, since mRNA
is much less stable than DNA. The mRNA of
eukaryotes is polyadenylated, meaning a poly(A)
tail is added to the 3' end of the final
transcript. Therefore, a primer consisting of
multiple 'T's can be made that will complimentary
base pair with the poly(A) tail of every mRNAs in
a cell. Once the primer has bound to the mRNA,
the enzyme reverse transcriptase can make a DNA
strand that is complimentary to the RNA. This DNA
strand will then be converted to a
double-stranded DNA molecule, which can then
proceed to the next step.
SAGE summary LongSAGE
Earliest versions 9 nt/tag Latest (LongSAGE)
20
Sequence get 15 tags per lane (300-400 bases)
1520 300
60 lanes per gel? 900 tags per gel 30 gels?
27000 tags
Figure 1. Differential gene expression is
responsible for the morphology of different
cells. All cells within an organism have the same
DNA, but not all genes are expressed. An
epithelial cell will express only genes specific
to skin, whereas a neuron will express genes
necessary for its development.
Painless Gene Expression ProfilingSAGE (Serial
Analysis of Gene Expression) Chan Ho Song
Michelle WyseGraphics Jiang Long With the
advent of the human genome project, a vast amount
of information about genes and gene structure is
suddenly at our fingertips. But this information
is limited. Every cell within an organism has the
same genetic composition (with the exception of
its gametes), and yet, obviously skin tissue is
very different from nervous tissue. The DNA
sequence cannot provide information about these
differences, which represent the next level of
complexity and organization within an organism
DNA expression. Cells within a multicellular
organism, such as ourselves, specialize to
perform specific functions to increase the
efficiency of the organism. Nerve cells, or
neurons, express neuron-specific proteins that
allow it to perform neuron duties. Skin, or
epithelial cells, have their own specific
proteins that enable their protective
functioning. Both neuron and epithelial cell have
the genes encoding for neural- and
epithelial-specific proteins, but each cell only
expresses the genes that it requires, and not
other tissue-specific genes (Figure 1). In this
way, a given DNA sequence only provides
information about what could be, not what
actually is.
9
(No Transcript)
10
Polonies
One primer is covalently incorporated into the
polyacrylamide
11
DAGE digital analysis of gene expression
One polony amplification at a time Counting
gives absolute frequency In mRNA population
Mikkilineni, V, Mitra, RD, Merritt, J, DiTonno,
JR, Church, GM, Ogunnaike, B, and Edwards, JS.
(2004) Digital Quantitative Measurements Of Gene
Expression. Biotechnology and Bioengineering
86(2)117-24.
12
Massively parallel DNA sequencing
Aqueous sphere in oil
Avidin Mag. Bead w primer
From shearing DNA
Template
Blunt end ligate
All spheres have PCR mix Productive spheres 1
bead, 1 template
Mme
Mme
Type IIS res. enz
Sequencing by ligation (see below)
Hybridization of amplified beads to low-density
capture beads (blue)
Millions of beads Embedded in polyacrylamide
Enrichment for amplified beads by centrifugation
Jay Shendure et al Science. (2005)
309(5741)1728-32 (2005)
13
Sequencing by ligation
Ligate
Anchor seq
Query seq
Query seq. for position 5 Cy54NA
5-Phos/NNNNANNNN/Cy5--3 Cy34NG
5-Phos/NNNNGNNNN/Cy3-3 TexasRed4NC
5-Phos/NNNNCNNNN/TR-3 FRET4NT
5-Phos/NNNNTNNNN/FRET-3 Preferential ligation
of no-mismatch hybrid.
262,144 nonamers degenerate at 8 of 9
positions Four pools for each position 6-7
positions for each genomic tag (of 8) Interrogate
both ends of each of the 2 tags 26 positions, 4
pools each, 104 pools total 26 cycles with
4-color probes Ligate, read color at each bead
position, strip Read colors ? computer Millions
of sequences in parallel
Use for gene expression?
14
Sequencing by ligation
15
(No Transcript)
16
SO4-2
APS adenosine phosphosulfate
17
(No Transcript)
18
(No Transcript)
19
Mammalian cell
genetics Introduction Genetics as a
subject (genetic things that go on in somatic
cells replicate, transmit, recombine, express
genes) Genetics as a tool. Best when you know
nothing. 4 manipulations of genetics 1-
Mutation in vivo (selection, usually) gene
knock-out or alteration in vitro site
directed or random cassette 2- Mapping
organismic mating ? recombination cell
culture cell fusion segregation radiation
hybrids FISH 3- Gene juxtaposition
(complementation) Organisms matings ?
heterozygotes Cell culture cell fusion ?
heterokaryons or hybrid cells 4- Gene transfer
transfection
20
Advantages of cultured cells numbers,
homogeneity Disadvantages of cultured mammalian
cells limited phenotypes limited
differentiation in culture (but some phenotypes
available) no sex (cf. yeast)
Mammalian cell lines Most genetic manipulations
use permanent lines, for the ability to do
multiple clonings Primary, secondary cultures,
passages, senescence. Crisis, established cell
lines, immortality vs. unregulated growth. Most
permanent lines immortalized, plus
"transformed, (plus have abnormal karyotypes)
21
Mutation in cultured mammalian cells Problem of
epigenetic change Variants vs. mutants Stable
heritable alterations in phenotype that are not
due to mutations heritable switches in
gene regulation (?) DNA CpG methylation,
histone acetylation/de-acetylation Diploidy.
Heteroploidy. Haploidy. The problem of diploidy
and heteroploidy (cf. e.g., yeast, or C. elegans,
Dros.) F2 ? homozygotes) Recessive mutations
(most knock outs) are masked.
22
Solutions to diploidy problem Dominant
mutations only (too limited) Haploid cells
(hasn't worked despite haploid frogs, and
modest chromosome reductions in CHO) Haploid
genes (XY) or functionally haploid (XX, allelic
exclusion)) Heterozygous loci (rare, despite
CHO reputation) Double mutants (incl. mutation
segregation, or mutation homozygosis(rare but
can be done) Heavy mutagenesis, Mutants/survivor
increases but mutants/ml decreases. How hard is
it to get mutants? What are the spontaneous and
induced mutation rates? Measurement of
spontaneous mutation rates. Rate vs. frequency.
Spont 10-7/cell-generation Induced
as high as 10-3 /cell (EMS, UV)
23
Loss of heterozygosity (LOH)by mitotic
recombination between homologous chromosomes
(rare)
-
-
1 homozygote 1 homozygote -
or
-
-
-
-
Heterozygote
After homologous recombination
-
-
-
2 heterozygotes again
24
Mutagenesis. Chemical and physical agents
MNNG point mutations (single base
substitutions) EMS
Bleomycin small
deletions UV mostly point mutations but also
large deletions Ionizing radiation (X-,
gamma-rays) large deletions, rearrangements Dosa
ge kill 90 Expression period dilute out
WT molecules (pre-existing protein. mRNA)
Metabolic cooperation WT toxic product can be
transferred.. Dominant vs. recessive mutations
Dom rare (subtle) but expressed, Recessives
easy but masked. Mutagen specificity (a
particular base or base combo). Mutational
spectra (hot and cold spots). Strand
specificity transcribed strand is often
preferentially repaired.
25
- Categories of cell mutants
- Exploitable metabolic pathways
- Purine and pyrimidine biosynthesis auxotrophs
- (auxotrophs require a nutrient in the medium that
the WT doesnt) - Amino acid biosynthesis auxotrophs
- Auxotrophs BUdR (BrdU) Kao and Puck. Kill
growing cells. Penicillin analogy. - Amino acid, nucleotide biosynthetic
pathways. - 2. Drug resistance see sheet
- A. Mutant lacks toxifying enzyme e.g., HPRT
(TGR), APRT (DAPR, 8-azaAR), TK (BrdUR) - B. Enzyme target becomes a better discriminator
(ouabain NaK ATPase pump) - C. Permeation changes influx blocked or efflux
increased. (MDR, P-glycoprotein) - D. Improved de-toxification via chelation,
covalent modification, - or overproduction of target (dhfr
MTX-resistance) - E. Receptor deficiency
- Glucocorticoid (steroid) receptor-resistant to
GC killing of lymphoid lines.
26
(No Transcript)
27
(No Transcript)
28
- 3. Temperature-sensitive mutants cell cycle
mutants. - Tritiated amino acid suicide (aa-tRNA
synthetases) - 4. Antibodies. Lysis with complement. Targets
cell surface constituents mostly (e.g., MHC) - 5. Visual inspection at colony level
- A. Sib selection (G6PD)
- B. Replica plating (LDH)
- C. Secreted product (Iganti-Ig IP)
- FACS fluorescence-activated cell sorter.
- 1-D and 2-D fluorescence displays (cell
surface Ag) - Brute force
- (clonal biochemical analysis, e.g.,
electrophoretic variants (e.g., Ig, isozymes) - Direct genotype analysis (rare) (DNA isolation
(via PCR and SSCP, single strand conformational
polymorphism electrophoresis. Or DGGE denaturing
gradient gel electrophoresis. - MHC major histocompatability locus or proteins
G6PD glucose-6-phosphate dehydrogenase LCH
lactate dehydrogenase Ig immunoglobulin
29
Cell fusion (for gene juxtaposition, protein
trafficking, mapping) Fusogenic agents PEG,
Sendai virus (syncytia promoting, as
HIV). Heterokaryons (2 nuclei), no cell
reproduction (limited times). (e.g., membrane
fluidity, nuclear shuttling, gene activation
(myoblasts) Hybrids (nuclei fuse, cells
reproduce). Complementation (e.g., auxotrophs
with same requirement) Dominance vs.
recessiveness. Mapping chromosome assignment.
Synteny. Radiation hybrids linkage analysis
30
Transfection agents DEAE-dextran (toxic, OK for
transient) CaPO4 (co-precipitate) Electroporation
(naked DNA, high quick voltage ? transient
holes) Lipofection (multilamellar
liposomes) Polybrene (detergent?) Ballistic
(DNA-coated gold particles) Must traverse
cytoplasm. Much engulfed in lysosomes.
Inhibition of lysosomal function often helps
(chloroquin) Pechelosome 2000 KB
co-integration of high MW DNA. Separate plasmids
-gt same site (co-integration). Separate
transfections -gt separate locations Random or
semi-random (many) integration sites (unless
targeted) Low but real homologous recombination
rate History discovered for practical use at
Columbia (PS Wigler Axel and Silverstein)
31
Transient transfection vs. permanent cloned
genes Transient -gt 10-50 transfection
efficiency (stain) Permanents more like 0.001
per µg DNA per cell (high). i.e., 106 -gt 1000
colonies could be much less for certain types of
cells
32
One the most dramatic first applications of gene
transfection from total DNA Transfer of the
growth-transformed phenotype ability to grow in
multilayers or in suspension in soft agar
(Weinberg, Wigler) DNA from tumor transfected
into growth controlled mouse 3T3 cells. Look
for foci (focus). Make a library from
growth-transformed transfectant. Screen for human
Alu repeat. Verify cloned DNA yields high
frequency of focus-forming transfectants. Isolate
cDNA by hybridization. Sequence. Identify gene
a dominant oncogene. Ras, a signaling protein
in transducing pathway for sensing growth factors
33
Recombination gene targeting Mitotic
recombination between homologous chromosomes
relation to cancer through the loss of tumor
suppressor genes LOH loss of homozygosity WT
/ ? mutation ? /- (WT phenotype) ? LOH
via homologous recombination, or loss and
duplication ? -/- Recombination of transfected
genes homologous vs. non-homologous
recombination. Gene conversion vs. reciprocal
recombination. Recombination between tandem
inserts (higher freq) Gene knockouts via
homologous recombination. ES cells and
transgenic mice. Selection for homologous
recombinants via the loss of viral TK genes
(Capecchi). Allele replacements in cultured cell
lines (e.g., APRT). Little work in cultured
lines myc double K.O. viable, sick
34
Gene amplification Historically Methotrexate
resistance (Littlefield) High dihydrofolate
reductase (DHFR) enzyme activity, protein,
protein synthetic rate, translatable mRNA.
(Schimke) mRNA level, DNA level. Homogeneously
staining, expanded chromosomal regions (HSRs)
Biedler Nunberg HSR dhfr genes. Double
minute chromosomes. Amplicons. Big (300 KB). Can
shrink, migrate.
35
HSR ? dmin upon DS break induced by a homing
endonuclease (I-SceI).
HSR homogeneously staining region Dmin double
minute chromosomes
Arnaud Coquelle, Lorène Rozier, Bernard
Dutrillaux and Michelle Debatisse ONCOGENE, 31
October 2002, Volume 21, Number 50, Pages
7671-7679 Induction of multiple double-strand
breaks within an hsr by meganucleaseI-SceI
expression or fragile site activation leads to
formation of double minutes and other chromosomal
rearrangements
36
Ampification models Over-replication, unequal
sister chromatid exchange, breakage and
fusion. In nature rDNA in oocytes,
Drosophila chorion genes. In medicine
chemotherapy resistance (MDR, P-glycoprotein)
cancer (myc, ras) In biotechnology high
level recombinant protein production in mammalian
cells