System Diagram PowerPoint PPT Presentation
1 / 1
Title: System Diagram
1
Planning Interventions For Gene Regulatory
Networks Daniel Bryce Seungchan Kim dan.bryce,
dolchan_at_asu.edu
Methods We test the feasibility of our
approach with the WNT5A gene regulatory network
first presented by Kim et al.1 and later used
for intervention problems by Datta et al.2.
WNT5A plays a significant role in the development
of melanoma and is known to induce the metastasis
of melanoma when highly expressed and its
regulatory influences are well studied 3. The
objective of our study is to use AI planning
techniques to devise intervention plan to reduce
the expression (down regulation) of WNT5A, so as
to inhibit metastasis. We use Boolean
functions to describe the gene regulatory
network, similar to 1,2. The predictor
functions define how each gene affects the
others, and allow us to derive a probabilistic
transition relation. Following Datta et al.2,
we formalize the WNT5A gene regulatory network as
a PBN 4 and study several planning problems by
adding intervention actions. Intervention actions
re-write the predictor functions of specific
genes to reflect, not how other genes predict the
next value of a target gene, but how an external
action sets the value. The approach taken by
Datta et al.2 finds intervention plans using a
finite horizon enumerative dynamic programming
formulation. By enumerative, we mean that they
generate every belief state that is reachable
from some finite length plan. In the dynamic
programming problems, the authors assign
differing rewards to intervention and
non-intervention actions, as well as negative
rewards to states where WNT5A is highly
regulated. The intervention plan minimizes
expected reward of acting and reaching
undesirable states. Our formulation is
similar to that of 2, but there are two
significant places where our approach differs. We
use a search algorithm instead of enumerative
dynamic programming, and retain a factored
representation of states and the transition
functions for different actions. Using a search
algorithm allows us to represent significantly
fewer belief states, and our factored
representation reduces the burden associated with
the plans we do explore. The search algorithm
works by generating the belief states reachable
after one step, then picking the best for
inclusion in the plan. The search continues by
only generating the belief states reachable by
the current plan. The search can revise its
current plan by backtracking over previous
choices. However, in practice search does not
generate every belief state, as would the work of
2. Results The result is that we can
search for longer plans and represent gene
regulatory values at a finer level of
discretization (we model either binary or ternary
expression levels for genes). Another practical
advantage of our approach is that we represent
the entire problem seamlessly with a logic based
language PPDDL 5. The computational leverage of
our approach is also encouraging.
Abstract Modeling the dynamics of cellular
processes has recently become a important
research area of many disciplines. One of the
most important reasons to model a cellular
process is to enable high-throughput in-silico
experiments that attempt to predict or intervene
in the process. These experiments can help
accelerate the design of therapies through their
cheap replication and alteration. While some
techniques exist for reasoning with cellular
processes, few take advantage of the flexible and
scalable algorithms popularized in AI research.
We apply AI planning techniques to a well-studied
gene regulatory network model and demonstrate its
clear advantage over existing methods. At a
broader level, we show where cellular process
models can fuel research for improved planning
technology.
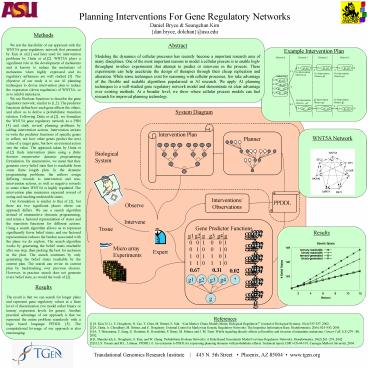
System Diagram
Biological System
Interventions/ Observations
Observe
Intervene
Gene Predictor Functions
Tissue
Micro array Experiments
Expert
0.67
0.31
0.02
g1
g2
g3
g4
?
g
References 1 S. Kim, H. Li, E. Dougherty, N.
Cao, Y. Chen, M. Bittner, E. Suh. Can Markov
Chain Models Mimic Biological Regulation?
Journal of Biological Systems, 10(4)337-357,
2002. 2 A. Datta, A. Choudhary, M. Bittner, and
E. Dougherty. External Control in Markovian
Genetic Regulatory Networks The Imperfect
Information Base. Bioinformatics, 20(6)924930,
2004. 3 A. T. Weeraratna, Y. Jiang, G.
Hostetter, K. Rosenblatt, P. Duray, M. Bittner,
and J. M. Trent. Wnt5a signaling directly affects
cell motility and invasion of metastatic
melanoma. Cancer Cell, 1(3)279 88, 2002. 4 I.
Shmulevich, E. Dougherty, S. Kim, and W. Zhang.
Probabilistic Boolean Networks A Rule-Based
Uncertainty Model For Gene Regulatory Networks.
Bioinformatics, 18(2)261274, 2002. 5 H.L.S.
Younes and M.L. Littman. PPDDL1.0 An extension
to PDDL for expressing planning domains with
probabilistic effects. Technical report,
CMU-CS-04-167, Carnegie Mellon University, 2004.