Abstract - PowerPoint PPT Presentation
1 / 1
Title:
Abstract
Description:
Regions with high rates of substitution or positive selection within the human ... that regions showing positive-selectivity across species substantiate a series ... – PowerPoint PPT presentation
Number of Views:75
Avg rating:3.0/5.0
Title: Abstract
1
Genes showing accelerated evolutionary properties
associate with language
K.J. Kelsey1, J.B. Tomblin2, J.B. Bjork1, S.K.
Iyengar3, L.E. Sucheston4, B.K. Samelson1, J.C.
Murray1 1) Dept. Pediatrics, The University of
Iowa, Iowa City, IA 2) Dept. Speech Pathology
and Audiology, University of Iowa, Iowa City, IA
3) Dept. Epidemiology and Biostatistics, Case
Western Reserve, Cleveland, OH 4) Dept.
Biostatistics, SUNY-Buffalo, Buffalo, NY.
Abstract
- Regions with high rates of substitution or
positive selection within the human genome are
potential candidates for studies including
language phenotypes. - Language phenotypes and DNA were collected on
triads of 581 second-grade children. - ASPM and the human accelerated region HAR1 were
typed using Taqman assays. - Two markers showed significant values for
various language measures in ASPM, and four
markers showed significant values around HAR1. - ASPM and the RNA genes, HAR1F and HAR1R, show
evidence of association for speech, language and
reading related measures.
Introduction
Results
Advances in comparative genomics have allowed for
the detection of evolutionarily-accelerated
regions within the human genome. The human
capacity to acquire complex language seems to be
unique, setting humans apart from other primates.
As such, it is hypothesized that regions showing
positive-selectivity across species substantiate
a series of candidate regions that may be
implicated in the study of human-specific traits,
such as speech, language, and reading. We
examined two positively-selected regions, the
gene ASPM and the human accelerated region (HAR)
HAR1, for association with a set of speech and
language related phenotypes under the hypothesis
that these behaviors would be under positive
selection. ASPM regulates brain size and is
implicated in neuronal stem cell proliferation
and differentiation. HAR1, a 118-bp region with
an estimated 18 substitutions in the human
lineage since human-chimpanzee divergence,
bisects two RNA genes, HAR1F and HAR1R. It forms
a stable structure and shows co-expression with
reelin, a regulator of neocortex development.
- ASPM
- Two markers, rs6700180 and rs10737686, showed
significant p-values across multiple measures
(see table 2 and figure 1) - HAR1
- Four markers, rs6011605, rs6089838, rs7506966,
and rs1884333, showed significant p-values across
multiple language measures (see table 2 and
figure 2) - Combined into a two SNP haplotype, rs6011605 and
rs6089838, show significant association with
spoken language (p.00003) - Sequencing revealed two novel SNPs within the
RNA genes HAR1F and HAR1R and next to HAR1 - chr2061204086 (MAF 0.33)
- chr2061204162 (MAF 0.24)
- Chr2061204086, between rs6011605 and rs750696,
only showed semi-significant association (p-value
0.08) for the Elision measure
ASPM
Table 2 Significant SNP results with associating
language measures
HAR1
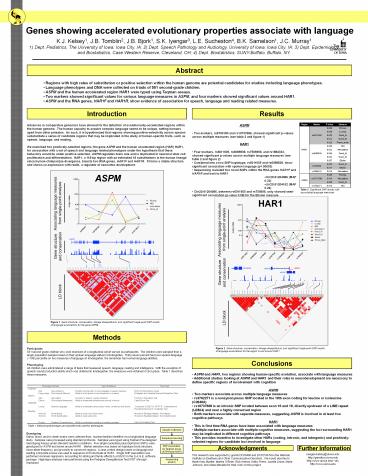
Figure 1 Gene structure, conservation, linkage
disequilibrium, and significant single-point SNP
results of language associations for the gene ASPM
Methods
Participants 581 second grade children who were
members of a longitudinal cohort served as
participants. The children were sampled from a
larger population sample based on their spoken
language status in kindergarten. Thirty-seven
percent had poor spoken language (lt10th
percentile on two measures of language) in
kindergarten, the remainder had normal language
abilities. Phenotyping All children were
administered a range of tasks that measured
speech, language, reading and intelligence. With
the exception of speech sound production ability
which was obtained in kindergarten, the measures
were obtained in 2nd grade. Table 1 describes
these measures.
Figure 2 Gene structure, conservation, linkage
disequilibrium, and significant single-point SNP
results of language associations for the region
in and around HAR1
Conclusions
- ASPM and HAR1, two regions showing
human-specific evolution, associate with language
measures - Additional studies looking at ASPM and HAR1 and
their roles in neurodevelopment are necessary to
define specific regions of involvement with
cognition - ASPM
- Two markers associate across multiple language
measures - rs3762271 is a nonsynonymous SNP located in the
18th exon coding for leucine or isoleucine
(C2648A) - rs10737686 is an intronic SNP located between
exon 18 and 19, directly upstream of a LINE
repeat (LIMA4) and near a highly conserved region - Both markers associate with separate measures,
suggesting ASPM is involved in at least two
cognitive pathways - HAR1
- This is first time RNA genes have been
associated with language measures - Multiple markers associate with multiple
cognitive measures, suggesting the loci
surrounding HAR1 may be implicated in different
cognitive pathways - This provides incentive to investigate other
HARs (coding, intronic, and intergenic) and
positively-selected regions for candidate loci
involved in language
Table 1. Behavioral phenotypes and specific tests
used for phenotypes.
Genotyping Saliva, blood, and/or cheek swabs were
collected from nuclear families identified via a
longitudinal language study. Samples were
processed using standard protocols. Samples were
typed using Taqman Pre-designed Genotyping Assays
under standard reaction conditions. Five single
nucleotide polymorphisms (SNPs) were genotyped
for ASPM and seven around HAR1. Marker selection
was based on haplotype block structure, minor
allele frequency, and proximity to conserved
regions. A case-control design based on language
and reading composite scores was used to sequence
218 individuals at HAR1. Single SNP association
was performed via linear regression, accounting
for sibling and family effects by ASSOC in the
S.A.G.E. software package. Haplotype analyses
were performed using the Pedigree Disequilibrium
Test (PDT) through Unphased.
Acknowledgments
Further Information
keegan-kelsey_at_uiowa.edu http//genetics.uiowa.edu
http//www.uiowa.edu/clrc http//www.uiowa.edu
This research was supported by grants DC00496 and
DC02746 from the National Institute on Deafness
and Other Communicative Disorders. We would also
like to thank Katy Mueller, Marlea OBrien, Rick
Arenas, Kathy Frees, Juanita Limas, Marla
Johnson, and Adela Mansilla for their work on
this project.