Project - PowerPoint PPT Presentation
Title:
Project
Description:
Project Studying Synechococcus elongatus for biophotovoltaics – PowerPoint PPT presentation
Number of Views:58
Avg rating:3.0/5.0
Title: Project
1
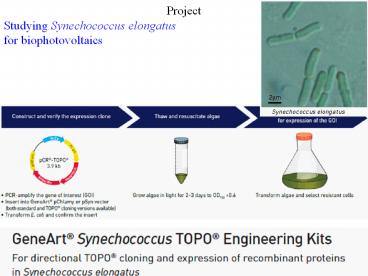
Project Studying Synechococcus elongatus for
biophotovoltaics
2
- How to bioengineer a novel bio-photovoltaic
system? - Obtain a sequence by PCR, then clone it into a
suitable plasmid - Were adding DNA, but want Synechococcus to make
a protein!
3
- In bacteria transcription and translation are
initially coupled
4
- In Bacteria transcription and translation are
initially coupled - RNA polymerase quits if ribosomes lag too much
5
- In Bacteria transcription and translation are
initially coupled - RNA polymerase quits if ribosomes lag too much
- Recent studies show that ribosomes continue
translating once mRNA is complete i.e after
transcription is done
6
- Bacteria have gt 1 protein/mRNA (polycistronic)
- http//bmb-it-services.bmb.psu.edu/bryant/lab/Proj
ect/Hydrogen/index.htmlsection1 - euk have 1 protein/mRNA
7
- Bacteria have gt 1 protein/mRNA (polycistronic)
- Mutations can have polar effects mutations in
upstream genes may affect expression of perfectly
good downstream genes!
8
Transcription Prokaryotes have one RNA
polymerase makes all RNA core polymerase
complex of 5 subunits (a1aIIbbw)
9
Transcription Prokaryotes have one RNA
polymerase makes all RNA core polymerase
complex of 5 subunits (a1aIIbbw) w not
absolutely needed, but cells lacking w are very
sick
10
Initiating transcription in Prokaryotes 1) Core
RNA polymerase is promiscuous
11
- Initiating transcription in Prokaryotes
- Core RNA polymerase is promiscuous
- sigma factors provide specificity
12
- Initiating transcription in Prokaryotes
- Core RNA polymerase is promiscuous
- sigma factors provide specificity
- Bind promoters
13
- Initiating transcription in Prokaryotes
- Core RNA polymerase is promiscuous
- sigma factors provide specificity
- Bind promoters
- Different sigmas bind different promoters
14
- Initiating transcription in Prokaryotes
- Core RNA polymerase is promiscuous
- sigma factors provide specificity
- Bind promoters
- 3) Once bound, RNA polymerase
- melts the DNA
15
- Initiating transcription in Prokaryotes
- 3) Once bound, RNA polymerase
- melts the DNA
- 4) rNTPs bind template
16
- Initiating transcription in Prokaryotes
- 3) Once bound, RNA polymerase
- melts the DNA
- 4) rNTPs bind template
- 5) RNA polymerase catalyzes phosphodiester bonds,
melts and unwinds template
17
- Initiating transcription in Prokaryotes
- 3) Once bound, RNA polymerase
- melts the DNA
- 4) rNTPs bind template
- 5) RNA polymerase catalyzes phosphodiester bonds,
melts and unwinds template - 6) sigma falls off after 10 bases are added
18
Structure of Prokaryotic promoters Three DNA
sequences (core regions) 1) Pribnow box at -10
(10 bp 5 to transcription start) 5-TATAAT-3
determines exact start site bound by s factor
19
Structure of Prokaryotic promoters Three DNA
sequences (core regions) 1) Pribnow box at -10
(10 bp 5 to transcription start) 5-TATAAT-3
determines exact start site bound by s
factor 2) -35 region 5-TTGACA-3 bound by
s factor
20
Structure of Prokaryotic promoters Three DNA
sequences (core regions) 1) Pribnow box at -10
(10 bp 5 to transcription start) 5-TATAAT-3
determines exact start site bound by s
factor 2) -35 region 5-TTGACA-3 bound by
s factor 3) UP element -57 bound by a factor
21
Structure of Prokaryotic promoters Three DNA
sequences (core regions) 1) Pribnow box at -10
(10 bp 5 to transcription start) 5-TATAAT-3
determines exact start site bound by s
factor 2) -35 region 5-TTGACA-3 bound by
s factor 3) UP element -57 bound by a factor
22
Structure of Prokaryotic promoters Three DNA
sequences (core regions) 1) Pribnow box at -10
(10 bp 5 to transcription start) 5-TATAAT-3
determines exact start site bound by s
factor 2) -35 region 5-TTGACA-3 bound by
s factor 3) UP element -57 bound by a
factor Other sequences also often influence
transcription! Eg CAP site in lac promoter
23
Structure of Prokaryotic promoters Other
sequences also often influence transcription! Our
plasmid contains the nickel promoter.
24
Structure of Prokaryotic promoters Other
sequences also often influence transcription! Our
plasmid contains the nickel promoter.
?
25
Structure of Prokaryotic promoters Other
sequences also often influence transcription! Our
plasmid contains the nickel promoter. nrsBACD
encode nickel transporters
26
- Structure of Prokaryotic promoters
- Other sequences also often influence
transcription! Our plasmid contains the nickel
promoter. - nrsBACD encode nickel transporters
- nrsRS encode two component signal transducers
- nrsS encodes a his kinase
- nrsR encodes a response regulator
27
- Structure of Prokaryotic promoters
- nrsRS encode two component signal transducers
- nrsS encodes a his kinase
- nrsR encodes a response regulator
- When nrsS binds Ni it kinases nrsR
28
- Structure of Prokaryotic promoters
- nrsRS encode two component signal transducers
- nrsS encodes a his kinase
- nrsR encodes a response regulator
- When nrsS binds Ni it kinases nrsR
- nrsR binds Ni promoter and activates
- transcription of both operons
29
Termination of transcription in prokaryotes 1)
Sometimes go until ribosomes fall too far behind
30
Termination of transcription in prokaryotes 1)
Sometimes go until ribosomes fall too far
behind 2) 50 of E.coli genes require a
termination factor called rho
31
Termination of transcription in prokaryotes 1)
Sometimes go until ribosomes fall too far
behind 2) 50 of E.coli genes require a
termination factor called rho 3) Our terminator
(rrnB) first forms an RNA hairpin, followed by an
8 base sequence TATCTGTT that halts transcription
32
Homologous recombination 1) DNA strands must be
capable of base-pairing
33
- Homologous recombination
- DNA strands must be capable of base-pairing
- DNA must get cut
34
- Homologous recombination
- DNA strands must be capable of base-pairing
- DNA must get cut
- Ends are processed to form single-strand overhangs
35
- Homologous recombination
- DNA strands must be capable of base-pairing
- DNA must get cut
- Ends are processed to form single-strand
overhangs - Single strand invades homolog (with help of RecA
protein)
36
- Homologous recombination
- DNA strands must be capable of base-pairing
- DNA must get cut
- Ends are processed to form single-strand
overhangs - Single strand invades homolog (with help of RecA
protein) - Must be capable of forming hybrid molecule!
37
- Homologous recombination
- DNA strands must be capable of base-pairing
- DNA must get cut
- Ends are processed to form single-strand
overhangs - Single strand invades homolog (with help of RecA
protein) - Must be capable of forming hybrid molecule!
- DNA polymerase adds on to end of invading molecule
38
- Homologous recombination
- DNA strands must be capable of base-pairing
- DNA must get cut
- Ends are processed to form single-strand
overhangs - Single strand invades homolog (with help of RecA
protein) forms Holliday junction - Branch migrates
39
- Homologous recombination
- DNA strands must be capable of base-pairing
- DNA must get cut
- Ends are processed to form single-strand
overhangs - Single strand invades homolog (with help of RecA
protein) forms Holliday junction - Branch migrates
- Holliday jn is
- cut DNA is ligated
40
- Homologous recombination
- DNA strands must be capable of base-pairing
- DNA must get cut
- Ends are processed to form single-strand
overhangs - Single strand invades homolog (with help of RecA
protein) forms Holliday junction - Branch migrates
- Holliday jn is
- cut DNA is ligated
- 7) Use mismatch repair
- to fix mismatches
41
- Homologous recombination
- DNA strands must be capable of base-pairing
- DNA must get cut
- Ends are processed to form single-strand
overhangs - Single strand invades homolog (with help of RecA
protein) forms Holliday junction - Branch migrates
- Holliday jn is
- cut DNA is ligated
- 7) Use mismatch repair
- to fix mismatches
- Why add selectable
- marker to new sequence
42
- Finding Orthologs
- Go to http//www.ncbi.nlm.nih.gov/
- Enter name of gene in search window
- Select nucleotide
- Select name of a promising sequence
- Select run BLAST optimize for somewhat similar
sequences (blastn) - Pick out interesting orthologs
43
- Finding Orthologs
- Go to http//www.ncbi.nlm.nih.gov/
- Select structure
- Enter name of protein in search window
- Select name of a promising sequence
- Select protein
- Select run BLAST
- Pick out interesting orthologs