Enzyme simulation PowerPoint PPT Presentation
Title: Enzyme simulation
1
Enzyme simulation
Concentration of substrate
Enzyme kinetics- Rate of enzyme catalysed
reaction is dependent upon
For an enzyme obeying michaelis menten
kinetics VVmaxS Km S
Vmax
V
Michaelis menten describes the initial rate of
reaction of a single substrate with enzyme under
steady state conditions.
S
Concentration of enzyme
The rate of reaction increases linearly as the
concentration of enzyme increases until all
substrate is utilized
Except when EgtgtS
V
E
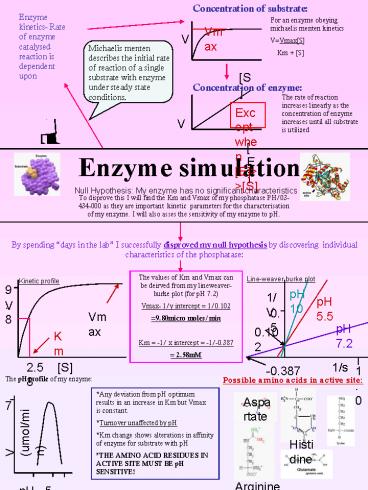
Null Hypothesis My enzyme has no significant
characteristics
To disprove this I will find the Km and Vmax of
my phosphatase PH/03-434-000 as they are
important kinetic parameters for the
characterisation of my enzyme. I will also asses
the sensitivity of my enzyme to pH.
By spending days in the lab I successfully
disproved my null hypothesis by discovering
individual characteristics of the phosphatase
The values of Km and Vmax can be deirved from my
lineweaver-burke plot (for pH 7.2) Vmax- 1/y
intercept 1/0.102 9.80micro moles/min Km
-1/ x intercept -1/-0.387 2.58mM
Line-weaver burke plot
Kinetic profile
9.8
pH 10
1/V
pH 5.5
V
0.5
Vmax
pH 7.2
0.102
Km
S
2.58
1/s
-0.387
1.0
The pH profile of my enzyme
Possible amino acids in active site
Any deviation from pH optimum results in an
increase in Km but Vmax is constant. Turnover
unaffected by pH Km change shows alterations in
affinity of enzyme for substrate with pH THE
AMINO ACID RESIDUES IN ACTIVE SITE MUST BE pH
SENSITIVE!
Aspartate
7
V (umol/min)
Histidine
Arginine
pH 5 7.2 10
PowerShow.com is a leading presentation sharing website. It has millions of presentations already uploaded and available with 1,000s more being uploaded by its users every day. Whatever your area of interest, here you’ll be able to find and view presentations you’ll love and possibly download. And, best of all, it is completely free and easy to use.
You might even have a presentation you’d like to share with others. If so, just upload it to PowerShow.com. We’ll convert it to an HTML5 slideshow that includes all the media types you’ve already added: audio, video, music, pictures, animations and transition effects. Then you can share it with your target audience as well as PowerShow.com’s millions of monthly visitors. And, again, it’s all free.
About the Developers
PowerShow.com is brought to you by CrystalGraphics, the award-winning developer and market-leading publisher of rich-media enhancement products for presentations. Our product offerings include millions of PowerPoint templates, diagrams, animated 3D characters and more.