In vitro systems have been used to investigate transcription' - PowerPoint PPT Presentation
1 / 19
Title:
In vitro systems have been used to investigate transcription'
Description:
TATA less promoters have a ... It consists of TATA box-binding protein (TBP) and eleven TBP-associated ... The TATA box and initiator are core ... – PowerPoint PPT presentation
Number of Views:30
Avg rating:3.0/5.0
Title: In vitro systems have been used to investigate transcription'
1
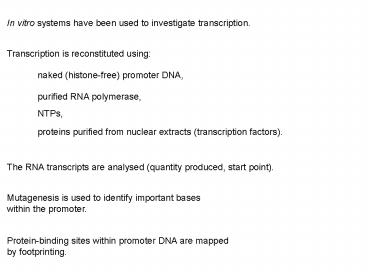
In vitro systems have been used to investigate
transcription.
Transcription is reconstituted using
naked (histone-free) promoter DNA,
purified RNA polymerase,
NTPs,
proteins purified from nuclear extracts
(transcription factors).
The RNA transcripts are analysed (quantity
produced, start point).
Mutagenesis is used to identify important bases
within the promoter.
Protein-binding sites within promoter DNA are
mapped by footprinting.
2
Pol II promoters
The minimal RNA pol II promoter has two key
features.
TATA box
Initiator Inr
TATA
YYCAYYYYY
-25
Start point
3
TATA less promoters have a downstream
positioning element (DPE) 28 to 32 nucleotides
downstream of the start point.
DPE
Initiator Inr
YYCAYYYYY
AGAC
30
4
Pol II initiation
RNA pol II requires several transcription factors
to initiate.
TFIID is an 800 kDa protein complex. It consists
of TATA box-binding protein (TBP) and eleven
TBP-associated protein factors (TAFs).
5
TATA box-binding protein
6
TBP sits on the TATA box like a saddle.
7
TBP binding widens the minor groove and bends
promoter DNA.
Purified TBP protects the region from -37 to -25.
Intact TFIID protects from - 45 to -10. Two of
the TAFs contact the start point.
8
TFIID is the only basal transcription factor that
makes sequence-specific contact with promoter
DNA.
Bound TFIID provides docking sites for other
components of the transcription apparatus.
The other TFIIs bind in a defined order A, B, F
RNA pol II, E, H and J.
9
TFIIB binds the bent face of DNA and protects the
region between -10 and 10.
10
TFIIA increases protection of the upstream region.
This complex assists binding of Pol II TFIIF
11
The initial complex of TFIID, TFIIA and TFIIB
recruits RNA pol II to the promoter and positions
it at the start point.
Polymerase binding covers DNA as far as 15 on
the template strand.
12
Then TFIIs E, H and J bind.
TFIIH
TFIIJ
TFIID
RNA pol II
TFIIA
TBP
TFIIF
TFIIB
TFIIE
TFIIE has helicase activity. TFIIH also has
helicase activity and a kinase activity
that phosphorylates the CTD of the largest pol II
subunit.
13
Phosphorylation releases Pol II from the basal
factors so that it can proceed to transcribe the
template.
Most of the TFIIs are released but TFIIH can
reassociate. A form of TFIIH may function in DNA
repair when RNA pol II is stalled by damage in
the template.
Some subunits of TFIIH are DNA repair proteins.
Absence of one of these proteins (through
mutation) results in the skin cancer Xeroderma
pigmentosum.
14
Patients with Xeroderma pigmentosum
15
The TATA box and initiator are core elements that
determine the start point and are sufficient for
low level transcription in vitro. Upstream
elements (UEs) are required for efficient
transcription in vivo. UEs bind activator
proteins that increase the efficiency of
transcription initiation.
16
Upstream elements - are located between -40 and
-110. - are generally quite short (10 bp). -
are orientation independent. - can have direct
or inverted repeats (transcription factors may
bind as dimers).
17
The CAAT box (consensus 5 GGNCAATCT 3) is
recognised by NF1 transcriptional
activators. The GC box (consensus 5 GGGCGG 3)
is another common element that binds a 60 kDa
activator protein Sp1.
-110
- 40
-25
Start point
CAAT box
GC box
TATA box
18
Factors bound to UEs contact basal factors TFIID,
TFIIB or TFIIA.
Protein-protein interactions favour assembly of
the transcription initiation complex.
19
Promoters containing just core and upstream
elements are active in all cell types. The
number of UEs determines the basal level of
transcription.