Fig. 7-0a - PowerPoint PPT Presentation
1 / 15
Title: Fig. 7-0a
1
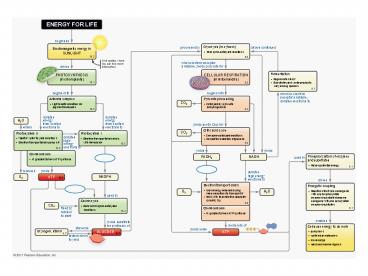
ENERGY FOR LIFE
begins as
Glycolysis (in cytosol)
processed by
allows continued
Electromagnetic energy in SUNLIGHT
10 enzyme-catalyzed reactions
10.2
9.3
Text section where you can find more information
when electron acceptor available, yields pyruvate
for
drives
PHOTOSYNTHESIS (in chloroplasts)
CELLULAR RESPIRATION (in mitochondria)
Fermentation
Regenerates NAD Substrates and waste
products vary among species
10.1
9.2
9.7
begins with
begins with
when no electron acceptor available, donates
electrons to
Antenna complex
Pyruvate processing
CO2
Light excites electrons in pigment
molecules
Catalyzed by pyruvate dehydrogenase
10.2
9.4
donates energy from excited electrons to
donates energy from excited electrons to
H2O
yields acetyl CoA for
enters
Citric acid cycle
CO2
Photosystem II
Photosystem I
8 enzyme-catalyzed reactions Completes
oxidation of glucose
donates high- energy electrons to
Splits water to yield electrons Electron
transport chain pumps H
Electron transport chain ends with
ferrodoxin
9.5
10.3
10.3
yields
yields
Chemiosmosis
NADH
FADH2
Phosphorylation of enzymes and substrates
used in
H gradient drives ATP synthase
Raises potential energy
9.1
releases
yields
donates high energy electrons to
O2
NADPH
ATP
9.1
drives
Electron transport chain
Energetic coupling
Uses energy released during redox
reactions to transport H Ends with final
electron acceptor (usually O2)
H2O
O2
Reactions that were endergonic with
unphosphorylated enzymes/substrates become
exergonic with phosphorylated
enzymes/subtrates
used in
Calvin cycle
9.6
CO2
Series of enzyme-catalyzed reactions
fixed by rubisco to start
Chemiosmosis
9.1
10.4
H gradient drives ATP synthase
enables
yields substrate for synthesis of
yields lots of
Cells use energy to do work
stored as
yields some
Glycogen, starch
GLUCOSE
pump ions synthesize molecules move
cargo send and receive signals
ATP
5.2
5.1
broken down to yield
2
ENERGY FOR LIFE
begins as
Electromagnetic energy in SUNLIGHT
10.2
Text section where you can find more information
drives
PHOTOSYNTHESIS (in chloroplasts)
10.1
begins with
Antenna complex
Light excites electrons in pigment molecules
10.2
donates energy from excited electrons to
donates energy from excited electrons to
H2O
enters
Photosystem II
Photosystem I
donates high- energy electrons to
Splits water to yield electrons Electron
transport chain pumps H
Electron transport chain ends with
ferrodoxin
10.3
10.3
Chemiosmosis
H gradient drives ATP synthase
releases
yields
O2
ATP
NADPH
9.1
used in
Calvin cycle
CO2
Series of enzyme-catalyzed reactions
fixed by rubisco to start
10.4
yields substrate for synthesis of
stored as
Glycogen, starch
GLUCOSE
5.2
5.1
broken down to yield
3
Glycolysis (in cytosol)
processed by
allows continued
10 enzyme-catalyzed reactions
9.3
when electron acceptor available, yields pyruvate
for
Fermentation
CELLULAR RESPIRATION (in mitochondria)
Regenerates NAD Substrates and waste
products vary among species
9.2
9.7
begins with
when no electron acceptor available, donates
electrons to
Pyruvate processing
CO2
Catalyzed by pyruvate dehydrogenase
9.4
yields acetyl CoA for
Citric acid cycle
CO2
8 enzyme-catalyzed reactions Completes
oxidation of glucose
9.5
yields
yields
NADH
FADH2
donates high energy electrons to
Electron transport chain
Uses energy released during redox reactions
to transport H Ends with final electron
acceptor (usually O2)
H2O
O2
9.6
Chemiosmosis
H gradient drives ATP synthase
yields lots of
yields some
ATP
4
Phosphorylation of enzymes and substrates
used in
ATP
9.1
Raises potential energy
9.1
drives
Energetic coupling
Reactions that were endergonic with
unphosphorylated enzymes/substrates become
exergonic with phosphorylated enzymes/subtrates
9.1
enables
Cells use energy to do work
pump ions synthesize molecules move
cargo send and receive signals
5
GENETIC INFORMATION
is archived in base sequences of
may change due to
DNA
Chromatin
Chromosomes
Breakage Duplication or deletion due
to errors in meiosis Damage by radiation
or other agents
4.2
18.2
11.1
Text section where you can find more information
is packaged with proteins to form
18.2
consists of functional units called
Genes
Genotype
Alleles
12.4
15.1
13.2
13.2
have different versions called
make up
14.5 15.4
can be
can be
are
causing
and
can be
EXPRESSED
COPIED
Mutation
TRANSMITTED
may regulate whether genes
15.4
15.2
14.3
11.1
can be
17.14 18.14
12.1, 13.14
if first TRANSCRIBED by
to somatic cells by
to germ cells by
by
Independent assortment Recombination
RNA polymerase
DNA polymerase
MITOSIS
MEIOSIS
11.1
12.1
includes
16.1
14.3
occasionally make errors, causing
starts with
starts with
to form
12.2 13.34
Parent cell
Parent cell
RNA
MUTATION
15.4
4.3
may be processed by
may function directly in cell as
2n
2n
Splicing Addition of 5? cap Addition of
poly(A) tail
ends with
ends with
16.2
tRNA (transfer RNA) rRNA (ribosomal
RNA)
16.4
n
n
to form
16.5
mRNA (messenger of RNA)
2n
2n
n
n
Two daughter cells with the same
genetic information as the parent cell
(unless mutation has occurred).
16.2
Four daughter cells with half the
genetic information as the parent cell.
is then TRANSLATED by
affect
Ribosomes
16.5
occurs during
occurs during
to form
GROWTH and ASEXUAL REPRODUCTION
SEXUAL REPRODUCTION
Proteins
3.2 16.5
11.0
12.3
changed by
result in
results in
Phenotype
Folding Glycosylation Phosphorylation
Degradation
3.4 5.3 9.1 18.4
13.1
Low genetic diversity
High genetic diversity
produce
6
GENETIC INFORMATION
is archived in base sequences of
DNA
4.2
Text section where you can find more information
consists of functional units called
Genotype
Genes
13.2
15.1
make up
can be
EXPRESSED
may regulate whether genes
15.2
17.14 18.14
if first TRANSCRIBED by
RNA polymerase
16.1
to form
RNA
4.3
7
RNA
4.3
may be processed by
may function directly in cell as
Splicing Addition of 5? cap Addition of
poly(A) tail
16.2
tRNA (transfer RNA) rRNA (ribosomal
RNA)
16.4
to form
16.5
mRNA (messeger of RNA)
16.2
is then TRANSLATED by
affect
Ribosomes
16.5
to form
Proteins
3.2 16.5
changed by
Folding Glycosylation Phosphorylation
Degradation
3.4 5.3 9.1 18.4
Phenotype
13.1
produce
8
may change due to
Breakage Duplication or deletion due to
errors in meiosis Damage by radiation or
other agents
Chromosomes
Chromatin
18.2
11.1
18.2
Alleles
12.4
13.2
14.5 15.4
are
can be
causing
and
can be
Mutation
TRANSMITTED
COPIED
15.4
14.3
11.1
can be
12.1, 13.14
to somatic cells by
to germ cells by
by
Independent assortment Recombination
DNA polymerase
MITOSIS
MEIOSIS
11.1
12.1
includes
14.3
occasionally make errors, causing
12.2 13.34
MUTATION
15.4
9
Independent assortment Recombination
MITOSIS
MEIOSIS
11.1
12.1
includes
starts with
starts with
12.2 13.34
Parent cell
Parent cell
2n
2n
ends with
ends with
n
n
2n
2n
n
n
Two daughter cells with the same
genetic information as the parent cell
(unless mutation has occurred).
Four daughter cells with half the
genetic information as the parent cell.
occurs during
occurs during
GROWTH and ASEXUAL REPRODUCTION
SEXUAL REPRODUCTION
11.0
12.3
result in
results in
Low genetic diversity
High genetic diversity
10
EVOLUTION
is
is
Change through time
Descent with modification
is
due to
due to
Changes in allele frequencies
25.1
does not produce
due to
due to
due to
due to
Inbreeding
Sexual selection
NATURAL SELECTION
GENETIC DRIFT
MUTATION
GENE FLOW
Mating among relatives Changes
genotype frequencies, but not allele
frequencies
Occurs when traits used in attracting
mates vary, and individuals with
certain traits attract the most mates
Occurs when traits vary, and individuals
with certain traits produce the most
offspring
Changes in allele frequencies due entirely
to chance Especially important in small
populations
Random changes in DNA Creates new alleles
Occurs in every individual in every
generation, at low frequency
Occurs when individuals move between
populations Homogenizes allele frequencies
between populations
includes
24.1 24.35 25.2
Gene flow
25.3
15.4, 25.5
25.4
due to lack of
exposes deleterious alleles to
25.6
25.6
produces divergence required for
includes
includes
produces divergence required for
produces divergence required for
Non-random mating
25.6
is the only evolutionary mechanism that can
produce
SPECIATION
The TREE OF LIFE
creates new branches on
Results from 1. Genetic isolation, followed
by 2. Genetic divergence
Text section where you can find more information
Describes the evolutionary
relationships among species
Adaptation
form smallest possible tips on
Involves heritable traits only
26.24
1.3, 27.1
prune
24.3, 24.5
forms new
MASS EXTINCTIONS
Species
60 of species are lost in less than 1
million years 5 events in the past 542
million years Is analogous to genetic drift
Evolutionarily independent units in nature,
identified by 1. Reproductive isolation,
and/or 2. Phylogenetic analysis, and/or 3.
Morphological differences
Fitness
26.1
27.4
Measured by number of offspring produced
may occur after
with
24.3, 24.5 25.16
Synamorphies
ADAPTIVE RADIATIONS
Rapid and extensive speciation in a single
lineage Dramatic divergence in morphology
or behavior (species use a wide array of
resources/habitats)
Traits that are unique to a single lineage
(found in some species but not others)
Arise in a common ancestor
that may be
may result in
Key innovations
usually reduces
Traits that allow species to exploit
resources in a new way or use new
habitats
26.1 27.1
27.3
27.4
11
EVOLUTION
is
is
Change through time
Descent with modification
is
due to
due to
Changes in allele frequencies
25.1
does not produce
due to
due to
due to
due to
Inbreeding
Sexual selection
NATURAL SELECTION
GENETIC DRIFT
MUTATION
GENE FLOW
Occurs when traits vary, and individuals
with certain traits produce the most
offspring
Changes in allele frequencies due
entirely to chance Especially important in
small populations
Occurs when individuals move between
populations Homogenizes allele frequencies
between populations
Mating among relatives Changes
genotype frequencies, but not allele
frequencies
Occurs when traits used in attracting
mates vary, and individuals with certain
traits attract the most mates
Random changes in DNA Creates new alleles
Occurs in every individual in every
generation, at low frequency
includes
24.1 24.35 25.2
Gene flow
25.3
15.4, 25.5
25.4
exposes deleterious alleles to
25.6
25.6
includes
includes
Non-random mating
25.6
is the only evolutionary mechanism that can
produce
Text section where you can find more information
Adaptation
Involves heritable traits only
24.3, 24.5
Fitness
Measured by number of offspring produced
24.3, 24.5 25.16
usually reduces
12
GENETIC DRIFT
MUTATION
GENE FLOW
NATURAL SELECTION
Changes in allele frequencies due entirely
to chance Especially important in small
populations
Random changes in DNA Creates new alleles
Occurs in every individual in every
generation, at low frequency
Occurs when traits vary, and individuals
with certain traits produce the most
offspring
Occurs when individuals move between
populations Homogenizes allele frequencies
between populations
24.1 24.35 25.2
Gene flow
25.3
15.4, 25.5
25.4
due to lack of
produces divergence required for
produces divergence required for
produces divergence required for
SPECIATION
The TREE OF LIFE
creates new branches on
Results from 1. Genetic isolation, followed
by 2. Genetic divergence
Describes the evolutionary
relationships among species
form smallest possible tips on
26.24
1.3, 27.1
prune
forms new
MASS EXTINCTIONS
Species
Evolutionarily independent units in nature,
identified by 1. Reproductive isolation,
and/or 2. Phylogenetic analysis, and/or 3.
Morphological differences
60 of species are lost in less than 1
million years 5 events in the past 542
million years Is analogous to genetic drift
27.4
26.1
may occur after
with
ADAPTIVE RADIATIONS
Synamorphies
Rapid and extensive speciation in a single
lineage Dramatic divergence in morphology
or behavior (species use a wide array of
resources/habitats)
Traits that are unique to a single lineage
(found in some species but not others)
Arise in a common ancestor
that may be
may result in
Key innovations
Traits that allow species to exploit
resources in a new way or use new
habitats
26.1 27.1
27.3
27.4
13
ECOLOGY
Text chapter or section where you can find more
information
is the study of how
interact with
Organisms
Abiotic environment
51
50.54
associate with others of the same species to form
includes
includes
includes
Populations
Energy
Water temperature Water flow rate Water
depth Nutrient availability
Soil Atmosphere
52
CO2
Chemical energy Solar energy
includes
50.34
and
Climate
54.1
Species
Communities
53.1
53
interact in
Temperature (especially average and
degree of yearly variation) Precipitation
(especially average and degree of yearly
variation)
and
Nutrients
interact via
form via
Carbon (C) Nitrogen (N) Phosphorous
(P) Others
Competition
Succession
?
?
/
Leads to possible exclusion of weaker
competitors Natural selection favors
traits that reduce competition
Pattern depends on species traits,
species interactions, and history of site
50.34
50.2, 54.3
54.2
53.2
dictates species that can be found in certain
53.1
is triggered by
flow through
influence
Terrestrial ecosystems
Aquatic ecosystems
Ecosystems
54
Disturbance
and
include
interact with abiotic factors to form
influence
50.3, 54.3
50.2, 54.3
E.g., fire, drought effect depends on
extent and frequency
Consumption (predation, parisitism, herbivory)
?
/
?
include
include
Primary producers (synthesize their own
food) Consumers (consume live organisms)
Decomposers (consume dead organisms)
53.3
Can reduce prey/host population size
Natural selection favors traits that
maximize defenses
affects
54.1
Species richness
53.4
53.1
form
affects
is a measure of
Primary productivity
Food webs
and
Biodiversity
Mutualism
flows through
/
?
?
Leads to co- occurrence of species
Natural selection favors traits that
maximize benefits and minimize costs
54.1
54.1
53.1
55
14
ECOLOGY
Text chapter or section where you can find more
information
is the study of how
interact with
Organisms
Abiotic environment
51
50, 54
associate with others of the same species to form
Populations
52
Species
Communities
53.1
53
Ecosystems
interact in
54
interact with abiotic factors to form
interact via
form via
Competition
Succession
?
?
/
Leads to possible exclusion of weaker
competitors Natural selection favors traits
that reduce competition
Pattern depends on species traits, species
interactions, and history of site
53.2
53.1
is triggered by
Disturbance
and
E.g., fire, drought effect depends on
extent and frequency
Consumption (predation, Parisitism, herbivory)
?
/
?
53.3
Can reduce prey/host population size
Natural selection favors traits that
maximize defenses
affects
Species richness
53.4
53.1
is a measure of
and
Biodiversity
Mutualism
/
?
?
Leads to co- occurrence of species
Natural selection favors traits that
maximize benefits and minimize costs
53.1
55
15
Abiotic environment
50.54
includes
includes
includes
Energy
Water temperature Water flow rate Water
depth Nutrient availability
Soil Atmosphere
CO2
Chemical energy Solar energy
includes
50.34
and
Climate
54.1
Temperature (especially average and
degree of yearly variation) Precipitation
(especially average and degree of yearly
variation)
and
Nutrients
Carbon (C) Nitrogen (N) Phosphorous
(P) Others
50.34
54.2
50.2, 54.3
dictates species that can be found in certain
flow through
influence
Aquatic ecosystems
Ecosystems
Terrestrial ecosystems
54
include
influence
50.3, 54.3
50.2, 54.3
include
include
Primary producers (synthesize their own
food) Consumers (consume live organisms)
Decomposers (consume dead organisms)
54.1
Species richness
53.4
form
affects
Primary productivity
Food webs
flows through
54.1
54.1