Folie 0 - PowerPoint PPT Presentation
1 / 1
Title: Folie 0
1
Stochastic Modelling of Single Cell Assay
Data S. Youssef, G. Schwake, S. Gude, J.O. Rädler
Department Physik and Center for NanoScience,
LMU München
Motivation Gene transfection, the introduction of
foreign genetic material into the nucleus of
eukaryotic cells, is of great importance for gene
technology in cell culture. Furthermore gene
delivery has therapeutic applications in medicine
as it enables the potential to treat several
previously incurable blood disorders and cancer
types. We use quantitative time-lapse
fluorescence microscopy to monitor the dynamics
of gene expression of large numbers of individual
cells in parallel. This approach reveals temporal
fluctuations and cell-to-cell variability that
would otherwise be masked by population-wide
measurements The goal is to establish computer
models for gene transfer that help to predict the
experimental outcome and hence will help to
design more efficient gene delivery agents. We
extend an implementation of the stochastic
Pi-calculus for our computer simulations 1. The
challenging task is to implement the topological
pathway as the gene carrier crosses many diverse
compartments and involves different steps in the
cytosolic transport.
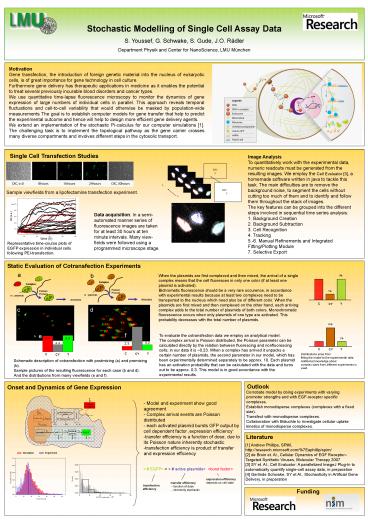
Single Cell Transfection Studies
Image Analysis To quantitatively work with the
experimental data, numeric readouts must be
generated from the resulting images. We employ
the Cell Evaluator 3, a homemade software
written in java to tackle this task. The main
difficulties are to remove the background noise,
to segment the cells without cutting too much of
them and to identify and follow them throughout
the stack of images. The key features can be
grouped into the different steps involved in
sequential time series analysis 1. Background
Creation 2. Background Subtraction 3. Cell
Recognition 4. Tracking 5.-6. Manual Refinements
and Integrated Fitting/Plotting Module 7.
Selective Export
Sample viewfields from a lipofectamine
transfection experiment.
Data acquisition In a semi-automated manner
series of fluorescence images are taken for at
least 30 hours at ten minute intervals. Many
view-fields were followed using a programmed
microscope stage.
Representative time-course plots of EGFP
expression in individual cells following
PEI-transfection.
Static Evaluation of Cotransfection Experiments
a
b
When the plasmids are first complexed and then
mixed, the arrival of a single complex means that
the cell fluoresces in only one color (if at
least one plasmid is activated). Bichromatic
fluorescence should be a very rare occurence, in
accordance with experimental results because at
least two complexes need to be transported to the
nucleus which need also be of different color.
When the plasmids are first mixed and then
complexed on the other hand, each arriving
complex adds to the total number of plasmids of
both colors. Monochromatic fluorescence occurs
when only plasmids of one type are activated.
This probability decreases with the total number
of plasmids.
9
1
C
Y
CY
d
c
14
f
e
To evaluate the cotransfection data we employ an
analytical model. The complex arrival is Poisson
distributed, the Poisson parameter can be
calculated directly by the relation between
fluorescing and nonfluorescing cells in our data
it is 0.23. When a complex has arrived it
unpacks a certain number of plasmids, the second
parameter in our model, which has been
experimentally determined separately to be
approx. 10. Each plasmid has an activation
probability that can be calculated with the data
and turns out to be approx. 0.3. This model is in
good accordance with the experimental results.
3
Distributions arise from fitting the model to
the experimental data. Additional knowledge
about complex sizes from different experiments is
used.
Schematic description of cotransfection with
postmixing (a) and premixing (b). Sample pictures
of the resulting fluorescence for each case (b
and d). And the distributions from many
viewfields (e and f).
Outlook
Onset and Dynamics of Gene Expression
Corrobate model by doing experiments with varying
promoter strengths and with EGF-receptor specific
complexes. Establish monodisperse complexes
(complexes with a fixed size). Transfect with
monodisperse complexes. Collaboration with
Bräuchle to investigate cellular uptake kinetics
of monodisperse complexes.
- Model and experiment show good agreement
- Complex arrival events are Poisson distributed
- each activated plasmid bursts GFP output by cell
dependent factor expression efficiency - transfer efficiency is a function of dose, due to
its Poisson nature inherently stochastic - transfection efficiency is product of transfer
and expression efficiency
Literature
1 Andrew Phillips, SPiM, http//research.microso
ft.com/7Eaphillip/spim/ 2 de Bruin et. Al.,
Cellular Dynamics of EGF ReceptorTargeted
Synthetic Viruses, Molecular Therapy 2007 3 SY
et. Al., Cell Evaluator A parallelized ImageJ
Plug-In to automatically quantify single-cell
assay data, in preparation 4 Gerlinde Schwake,
SY et Al., Stochasticity in Artificial Gene
Delivery, in preparation
Funding